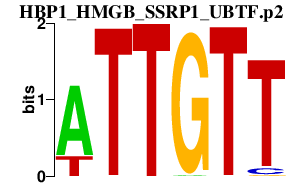
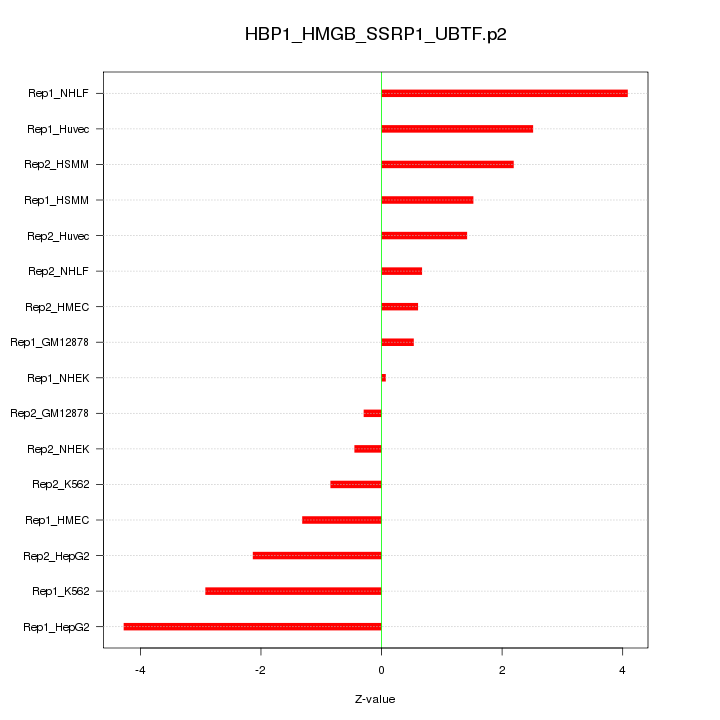
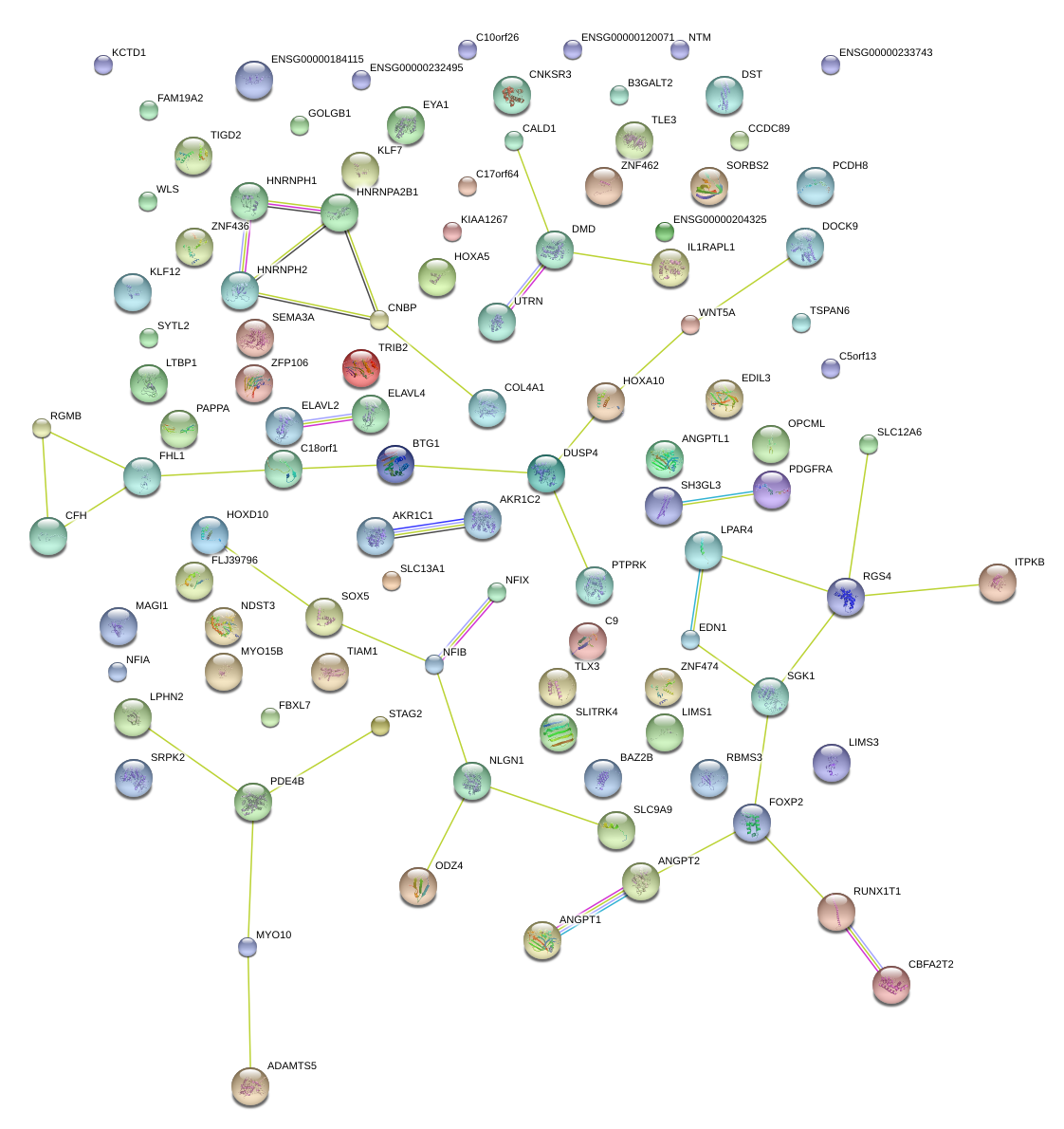
|
chr1_+_81544432
|
6.595
|
|
LPHN2
|
latrophilin 2
|
|
chrX_+_135079461
|
4.380
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr9_+_108665168
|
4.354
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr1_+_161305188
|
4.112
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr9_-_23816062
|
3.578
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_-_224993498
|
3.569
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr4_-_186814852
|
3.342
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_207738858
|
3.290
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr11_+_131286513
|
3.007
|
|
NTM
|
neurotrimin
|
|
chr8_-_6407882
|
2.961
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr7_-_104696678
|
2.909
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr7_-_27186400
|
2.880
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chrX_+_123062143
|
2.806
|
|
STAG2
|
stromal antigen 2
|
|
chr13_-_73605914
|
2.734
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr8_-_72436519
|
2.535
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr6_-_56824672
|
2.519
|
|
DST
|
dystonin
|
|
chr3_+_174598937
|
2.386
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr2_-_160181206
|
2.281
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr9_+_126460511
|
2.251
|
|
|
|
|
chr7_-_26202626
|
2.248
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr3_-_145049853
|
2.236
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chrX_+_28515479
|
2.223
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr5_+_15553736
|
2.183
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr10_+_104525877
|
2.183
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_-_65999548
|
2.176
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr7_-_27149750
|
2.143
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr3_-_130373241
|
2.137
|
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr3_+_148593911
|
2.127
|
|
|
|
|
chr13_-_59223074
|
2.122
|
|
|
|
|
chr6_-_134540666
|
2.110
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr4_+_119174947
|
2.096
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr8_-_93099040
|
2.065
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr18_-_22382497
|
2.039
|
NM_001142730
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr1_-_177106702
|
1.998
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr13_-_98428244
|
1.997
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr21_-_27261276
|
1.993
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr4_-_186815116
|
1.988
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_+_131285747
|
1.974
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr7_-_83662152
|
1.927
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr8_-_29262240
|
1.926
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr5_-_111119846
|
1.914
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr9_+_117955890
|
1.907
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr2_+_12775814
|
1.898
|
|
|
|
|
chr6_+_12398514
|
1.887
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chrX_+_77889861
|
1.872
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr11_-_85107569
|
1.871
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr3_+_29297806
|
1.865
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr19_+_12995782
|
1.853
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr8_-_108579270
|
1.829
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr1_+_66570363
|
1.780
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr5_-_88004891
|
1.775
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr6_-_56824371
|
1.774
|
|
DST
|
dystonin
|
|
chr17_-_41625921
|
1.750
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr9_-_14303509
|
1.741
|
|
NFIB
|
nuclear factor I/B
|
|
chr11_-_85108030
|
1.711
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr20_+_31613800
|
1.708
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr5_+_121493113
|
1.706
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr3_-_55496370
|
1.701
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr3_-_122951200
|
1.700
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chrX_+_135079120
|
1.698
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_-_33267646
|
1.669
|
NM_000109
|
DMD
|
dystrophin
|
|
chr17_+_71095733
|
1.656
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chrX_-_99778340
|
1.613
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr12_-_24606616
|
1.606
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr17_+_55854636
|
1.600
|
NM_181707
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr15_-_68177244
|
1.586
|
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr4_+_54790190
|
1.573
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr7_+_134114912
|
1.572
|
|
CALD1
|
caldesmon 1
|
|
chr21_-_31853160
|
1.567
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr5_-_178977441
|
1.566
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr2_+_12775336
|
1.555
|
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr4_+_90252990
|
1.548
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr7_-_122627235
|
1.519
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr5_+_98132898
|
1.512
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr1_+_161305558
|
1.508
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_-_55496607
|
1.502
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr6_-_128883125
|
1.501
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr20_+_56369012
|
1.499
|
|
|
|
|
chr7_+_134114957
|
1.495
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_191422346
|
1.490
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr1_-_68470585
|
1.486
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr13_-_73606362
|
1.481
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr15_-_68177280
|
1.478
|
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr5_+_140730014
|
1.474
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chrX_-_142551587
|
1.454
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr13_-_52320774
|
1.449
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr5_+_170668892
|
1.418
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr15_-_68177283
|
1.417
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr5_-_16989371
|
1.416
|
NM_012334
|
MYO10
|
myosin X
|
|
chr2_-_110665188
|
1.404
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr8_-_93184629
|
1.395
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_23568854
|
1.385
|
NM_001077195
|
ZNF436
|
zinc finger protein 436
|
|
chr12_-_91063441
|
1.384
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr7_+_113853790
|
1.372
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr3_-_194118321
|
1.365
|
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr11_-_78829342
|
1.364
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr15_+_81907176
|
1.359
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr15_-_32398220
|
1.347
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr2_+_33213111
|
1.337
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr18_+_13601664
|
1.324
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr11_-_85074848
|
1.312
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr5_-_71838875
|
1.312
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr5_-_83716346
|
1.310
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr13_-_109757442
|
1.303
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr5_-_39400319
|
1.301
|
NM_001737
|
C9
|
complement component 9
|
|
chr4_-_41579325
|
1.295
|
|
|
|
|
chr18_-_13905534
|
1.294
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr5_+_15553768
|
1.292
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr6_+_21774624
|
1.277
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr1_+_168898898
|
1.270
|
|
PRRX1
|
paired related homeobox 1
|
|
chr1_+_22107137
|
1.269
|
|
RPL21
|
ribosomal protein L21
|
|
chr9_+_27099138
|
1.258
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr1_+_148306439
|
1.256
|
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae)
|
|
chr3_-_69254226
|
1.246
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr22_+_36422940
|
1.244
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr13_+_23742846
|
1.236
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr5_-_146813343
|
1.229
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr9_+_115678365
|
1.224
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr4_+_37655385
|
1.220
|
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr3_+_116824840
|
1.216
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr1_+_84540921
|
1.212
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr4_+_3264509
|
1.210
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr5_-_39460687
|
1.206
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_+_13240983
|
1.201
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr8_-_72437020
|
1.199
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr11_-_69228286
|
1.197
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr6_+_99979490
|
1.194
|
|
|
|
|
chr8_-_72436832
|
1.187
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr4_-_111777651
|
1.185
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr12_+_14429499
|
1.182
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr1_+_24158898
|
1.177
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr7_+_28692122
|
1.174
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_+_13240987
|
1.167
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr7_-_27171673
|
1.159
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr5_+_15553539
|
1.159
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr18_-_21184824
|
1.149
|
|
|
|
|
chr5_+_148766632
|
1.146
|
|
LOC728264
|
hypothetical LOC728264
|
|
chr7_-_27108826
|
1.145
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr10_+_80672842
|
1.143
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr6_-_134680888
|
1.142
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_-_83715947
|
1.134
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr7_+_77005287
|
1.130
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr12_-_88270375
|
1.124
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr3_-_11585264
|
1.120
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_24158887
|
1.118
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr7_-_44854188
|
1.111
|
|
H2AFV
|
H2A histone family, member V
|
|
chr9_-_14303899
|
1.108
|
|
NFIB
|
nuclear factor I/B
|
|
chr3_-_115960807
|
1.107
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr13_-_35327798
|
1.099
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr7_-_44854222
|
1.094
|
NM_012412
NM_138635
NM_201436
NM_201516
NM_201517
|
H2AFV
|
H2A histone family, member V
|
|
chr14_+_85066265
|
1.091
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chrX_-_138742050
|
1.086
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr2_+_110013297
|
1.086
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr7_-_11838260
|
1.086
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr2_-_56266408
|
1.086
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr7_-_44854183
|
1.084
|
|
H2AFV
|
H2A histone family, member V
|
|
chr7_-_44854145
|
1.076
|
|
H2AFV
|
H2A histone family, member V
|
|
chr3_-_129023850
|
1.074
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr8_+_70567634
|
1.068
|
|
SULF1
|
sulfatase 1
|
|
chr5_+_140719773
|
1.067
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr8_-_82557956
|
1.067
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr20_+_11846536
|
1.067
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr4_-_83569853
|
1.063
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr3_-_66634031
|
1.061
|
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr12_+_47498778
|
1.049
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr16_+_31391998
|
1.048
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr1_+_196874867
|
1.047
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_154120170
|
1.047
|
|
SYT11
|
synaptotagmin XI
|
|
chr5_-_114543632
|
1.044
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr7_-_47588652
|
1.043
|
|
TNS3
|
tensin 3
|
|
chr2_-_121875610
|
1.036
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr5_+_61910318
|
1.034
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr15_+_81906983
|
1.027
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr4_-_138673078
|
1.020
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr19_+_60858230
|
1.016
|
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chr22_+_36928972
|
1.015
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr9_-_97308687
|
1.011
|
|
PTCH1
|
patched 1
|
|
chr10_+_119292699
|
1.011
|
|
|
|
|
chr5_-_39461054
|
1.006
|
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr11_+_129496792
|
1.006
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr8_+_70567581
|
1.005
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr4_-_176970372
|
1.000
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_+_39874921
|
0.994
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr9_-_14304036
|
0.989
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr7_-_44854092
|
0.988
|
|
H2AFV
|
H2A histone family, member V
|
|
chr2_+_108637698
|
0.987
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_+_36777336
|
0.986
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr4_+_24923490
|
0.985
|
NM_024936
|
ZCCHC4
|
zinc finger, CCHC domain containing 4
|
|
chr12_-_16649211
|
0.981
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_+_78297418
|
0.979
|
|
CCNG2
|
cyclin G2
|
|
chr9_-_72673753
|
0.977
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr4_+_183302105
|
0.975
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr9_-_16717887
|
0.973
|
|
BNC2
|
basonuclin 2
|
|
chr17_-_41626032
|
0.972
|
NM_015443
|
KIAA1267
|
KIAA1267
|
|
chr11_+_19328846
|
0.964
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr7_+_18501876
|
0.955
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr3_+_180348818
|
0.955
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr7_-_47588266
|
0.954
|
|
TNS3
|
tensin 3
|
|
chr9_-_71476917
|
0.950
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr3_-_48576204
|
0.950
|
NM_033199
|
UCN2
|
urocortin 2
|