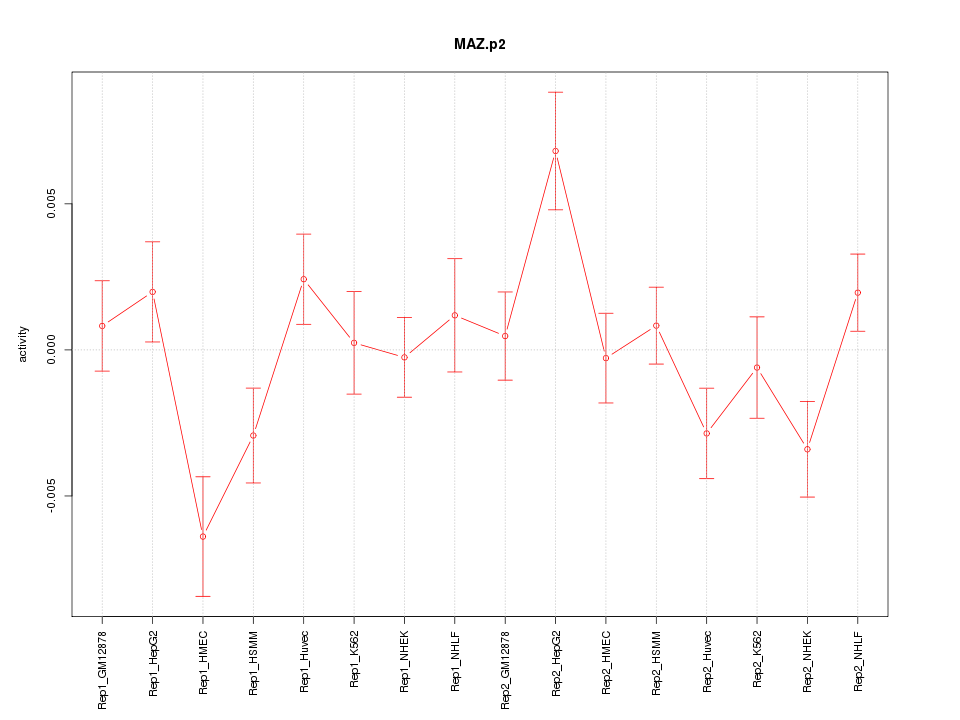
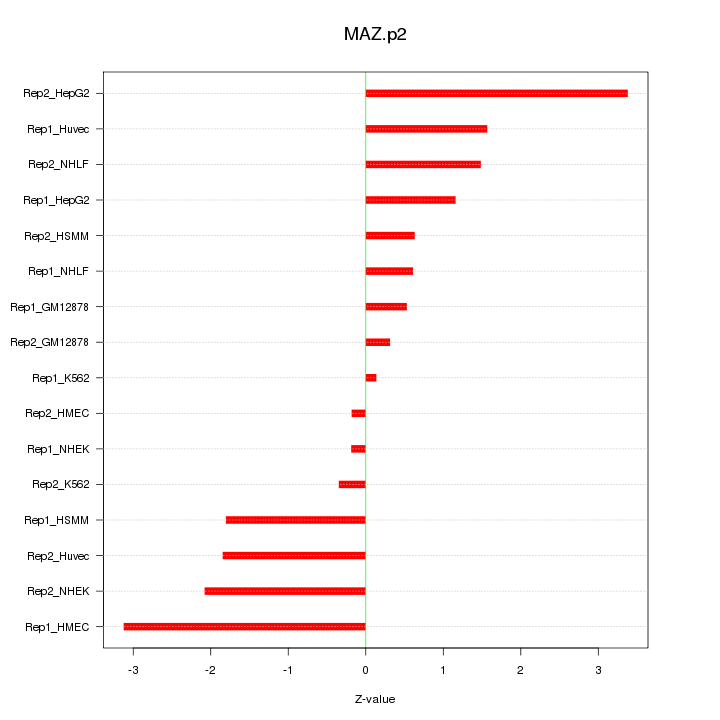
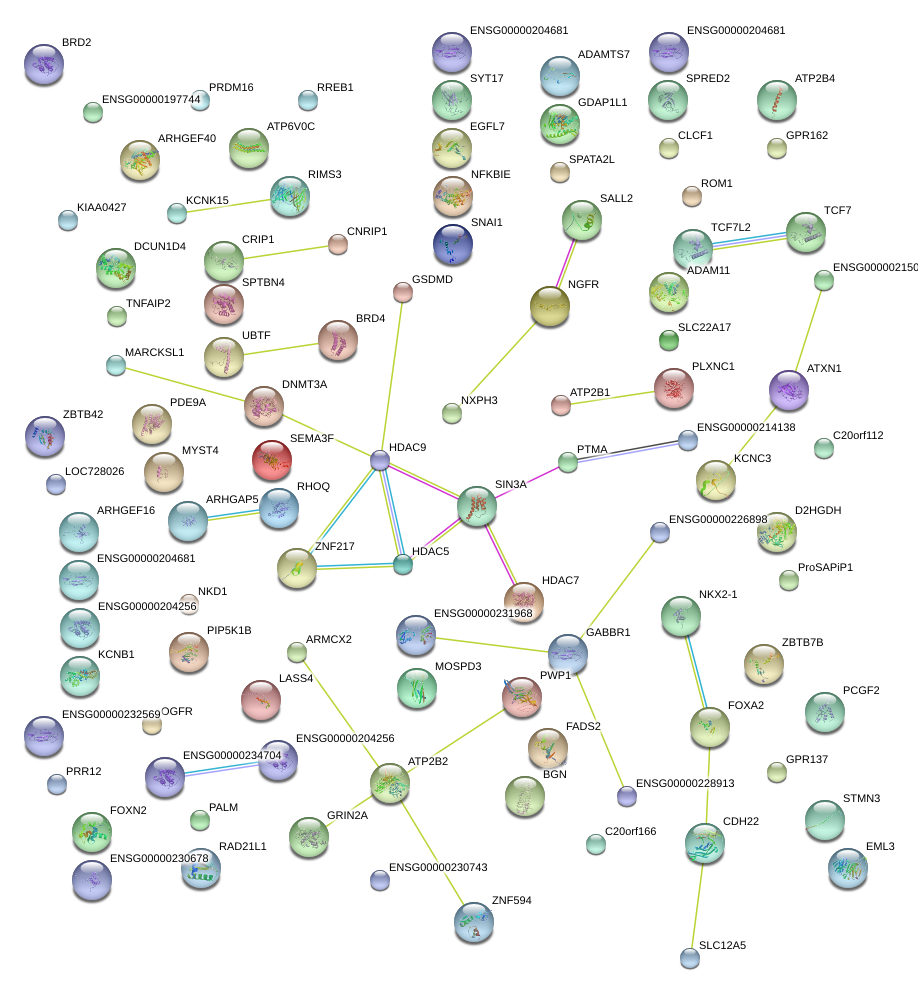
|
chr20_-_51644187
|
2.632
|
|
ZNF217
|
zinc finger protein 217
|
|
chr6_-_29703725
|
1.901
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr21_+_42946719
|
1.807
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr20_+_1154763
|
1.783
|
NM_001136566
|
RAD21L1
|
RAD21-like 1 (S. pombe)
|
|
chr17_-_34157962
|
1.694
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chr16_+_49139694
|
1.651
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr17_+_45008473
|
1.628
|
|
NXPH3
|
neurexophilin 3
|
|
chr1_-_40903897
|
1.534
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr6_-_29703913
|
1.525
|
NM_021903
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr2_+_46623325
|
1.515
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr1_-_40903700
|
1.497
|
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr19_-_55524445
|
1.463
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr1_-_32574185
|
1.410
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr17_+_45008175
|
1.324
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr14_+_31615922
|
1.320
|
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr16_+_19087035
|
1.294
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr9_+_70509925
|
1.288
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr14_+_20608179
|
1.275
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr10_+_76256305
|
1.258
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr20_-_44370537
|
1.253
|
|
CDH22
|
cadherin 22, type 2
|
|
chr8_+_144706699
|
1.250
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr20_+_42807901
|
1.240
|
NM_022358
|
KCNK15
|
potassium channel, subfamily K, member 15
|
|
chr6_-_16869579
|
1.230
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr20_-_3097070
|
1.207
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr14_+_31615959
|
1.201
|
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr14_-_36059084
|
1.194
|
NM_001079668
|
NKX2-1
|
NK2 homeobox 1
|
|
chr12_+_93066370
|
1.191
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr6_+_7052798
|
1.172
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr14_+_104337923
|
1.156
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr3_-_10522267
|
1.154
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr11_-_66897584
|
1.149
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr19_+_659766
|
1.147
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr3_-_10522221
|
1.147
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr2_+_46623396
|
1.141
|
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr6_+_33046706
|
1.138
|
|
BRD2
|
bromodomain containing 2
|
|
chr20_-_61755145
|
1.061
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr11_-_62136778
|
1.051
|
NM_153265
|
EML3
|
echinoderm microtubule associated protein like 3
|
|
chr1_+_153241858
|
1.041
|
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr16_+_2503676
|
1.039
|
NM_001198569
NM_001694
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr18_+_44319365
|
1.035
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr20_+_60906605
|
1.035
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr10_+_114699953
|
1.030
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr19_+_45728270
|
1.028
|
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr6_+_33046636
|
1.015
|
NM_001113182
|
BRD2
|
bromodomain containing 2
|
|
chr17_-_39556483
|
1.004
|
|
HDAC5
|
histone deacetylase 5
|
|
chr2_-_25328452
|
1.003
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr7_+_100047877
|
0.990
|
NM_001040098
NM_001040099
NM_023948
|
MOSPD3
|
motile sperm domain containing 3
|
|
chr12_-_46499834
|
0.987
|
NM_001098416
NM_015401
|
HDAC7
|
histone deacetylase 7
|
|
chr17_-_39652449
|
0.961
|
NM_001076684
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr9_+_138680064
|
0.958
|
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr17_+_40192072
|
0.955
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr14_+_31616196
|
0.954
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr11_+_61352348
|
0.949
|
|
FADS2
|
fatty acid desaturase 2
|
|
chr20_+_42309371
|
0.923
|
|
GDAP1L1
|
ganglioside-induced differentiation-associated protein 1-like 1
|
|
chr20_-_22512893
|
0.915
|
|
FOXA2
|
forkhead box A2
|
|
chr20_+_60558104
|
0.908
|
NM_178463
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr14_+_102659464
|
0.907
|
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr16_-_88295595
|
0.900
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr4_+_52403922
|
0.895
|
NM_001040402
NM_015115
|
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr17_-_39556535
|
0.895
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr1_+_153241665
|
0.889
|
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr19_+_8180179
|
0.886
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr3_-_10522340
|
0.885
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr2_+_242322645
|
0.884
|
NM_152783
|
D2HGDH
|
D-2-hydroxyglutarate dehydrogenase
|
|
chr16_-_10183244
|
0.877
|
NM_001134408
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr20_-_47532585
|
0.870
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr11_+_62136788
|
0.864
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr6_-_44341222
|
0.860
|
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr14_-_21064182
|
0.854
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr11_+_63809877
|
0.845
|
|
GPR137
|
G protein-coupled receptor 137
|
|
chrX_+_152413540
|
0.845
|
NM_001711
|
BGN
|
biglycan
|
|
chr19_+_45728207
|
0.845
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr15_-_76890626
|
0.844
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr6_+_7052985
|
0.843
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr2_-_65513138
|
0.841
|
NM_181784
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr14_-_22891865
|
0.837
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr11_+_63810157
|
0.834
|
NM_001170880
NM_020155
|
GPR137
|
G protein-coupled receptor 137
|
|
chr20_+_44083667
|
0.832
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr17_-_39652391
|
0.822
|
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr20_-_30534848
|
0.813
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr20_+_48032918
|
0.808
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr2_+_48395271
|
0.807
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr17_+_44927591
|
0.804
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr16_+_2503915
|
0.796
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr16_+_2504001
|
0.795
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr15_-_73530829
|
0.795
|
NM_015477
|
SIN3A
|
SIN3 homolog A, transcription regulator (yeast)
|
|
chr12_+_6808025
|
0.791
|
|
LEPREL2
|
leprecan-like 2
|
|
chr3_+_50167424
|
0.789
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr14_+_105024301
|
0.785
|
NM_001311
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr19_+_54786723
|
0.782
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr1_+_2975590
|
0.779
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr2_+_232281470
|
0.774
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr1_+_3378030
|
0.769
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr17_-_24918052
|
0.768
|
NM_198147
|
ABHD15
|
abhydrolase domain containing 15
|
|
chr6_-_33268177
|
0.768
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr17_+_78630793
|
0.765
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr20_-_17610828
|
0.762
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr20_+_43996717
|
0.753
|
NM_022104
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1
|
|
chr2_+_62786290
|
0.752
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr20_-_49592444
|
0.750
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr12_+_56230110
|
0.743
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr17_-_7095622
|
0.736
|
NM_001143775
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chr7_-_4889772
|
0.735
|
NM_018059
|
RADIL
|
Ras association and DIL domains
|
|
chr20_+_55359551
|
0.731
|
NM_003610
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr12_-_46438447
|
0.731
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr20_+_17498507
|
0.731
|
NM_001011546
NM_006870
|
DSTN
|
destrin (actin depolymerizing factor)
|
|
chr20_+_56900116
|
0.730
|
NM_001077488
|
GNAS
|
GNAS complex locus
|
|
chr1_+_3360880
|
0.725
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr20_-_49592654
|
0.723
|
NM_012340
NM_173091
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr1_+_210525490
|
0.721
|
NM_006243
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr17_-_70367479
|
0.719
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr6_+_33495824
|
0.717
|
NM_006772
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr1_+_45542138
|
0.715
|
|
LOC400752
|
hypothetical LOC400752
|
|
chr1_+_26671548
|
0.713
|
|
HMGN2
|
high-mobility group nucleosomal binding domain 2
|
|
chr10_+_98582648
|
0.712
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr17_-_38081967
|
0.707
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr7_-_100262971
|
0.706
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr19_+_59061422
|
0.702
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr20_+_34603311
|
0.702
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr17_+_7195931
|
0.700
|
NM_001002914
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr2_-_20288108
|
0.700
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr2_+_46779553
|
0.698
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chrX_+_152413646
|
0.695
|
|
BGN
|
biglycan
|
|
chr2_+_24153255
|
0.692
|
|
LOC375190
|
hypothetical protein LOC375190
|
|
chr17_-_7095982
|
0.691
|
NM_015343
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chr1_+_3679168
|
0.691
|
NM_001163724
|
LOC388588
|
hypothetical protein LOC388588
|
|
chr14_+_23508922
|
0.689
|
NM_001193636
NM_001193637
NM_001193635
|
DHRS4L2
DHRS4L1
|
dehydrogenase/reductase (SDR family) member 4 like 2
dehydrogenase/reductase (SDR family) member 4 like 1
|
|
chr12_+_6745947
|
0.687
|
|
PTMS
|
parathymosin
|
|
chr12_+_6745835
|
0.686
|
|
PTMS
|
parathymosin
|
|
chr13_+_75108450
|
0.683
|
|
LMO7
|
LIM domain 7
|
|
chr13_+_99431417
|
0.680
|
|
|
|
|
chr1_-_23683214
|
0.679
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr19_-_54636425
|
0.679
|
NM_020309
|
SLC17A7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7
|
|
chr14_+_23610582
|
0.678
|
NM_006032
|
CPNE6
|
copine VI (neuronal)
|
|
chr11_-_65573226
|
0.677
|
NM_033036
|
GAL3ST3
|
galactose-3-O-sulfotransferase 3
|
|
chr16_-_70475381
|
0.673
|
|
ZNF821
|
zinc finger protein 821
|
|
chr21_+_44699796
|
0.667
|
NM_030891
|
LRRC3
|
leucine rich repeat containing 3
|
|
chr2_-_148494732
|
0.662
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr18_-_28606839
|
0.660
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr20_-_61600834
|
0.653
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr11_+_64705855
|
0.653
|
NM_001198870
NM_005186
|
CAPN1
|
calpain 1, (mu/I) large subunit
|
|
chr2_+_232281491
|
0.648
|
|
PTMA
|
prothymosin, alpha
|
|
chr16_-_8870343
|
0.643
|
NM_014316
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr20_+_48240705
|
0.642
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr1_-_39910296
|
0.640
|
NM_032526
|
NT5C1A
|
5'-nucleotidase, cytosolic IA
|
|
chr19_-_7896974
|
0.638
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr22_-_40652724
|
0.637
|
NM_052945
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C
|
|
chr20_+_42309357
|
0.637
|
|
GDAP1L1
|
ganglioside-induced differentiation-associated protein 1-like 1
|
|
chr1_+_39319675
|
0.636
|
NM_012090
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr8_+_22078589
|
0.635
|
NM_001199
NM_006129
|
BMP1
|
bone morphogenetic protein 1
|
|
chr1_-_226661089
|
0.635
|
NM_145214
|
TRIM11
|
tripartite motif containing 11
|
|
chr11_+_64705967
|
0.635
|
|
CAPN1
|
calpain 1, (mu/I) large subunit
|
|
chr17_+_16259812
|
0.634
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr2_-_60634271
|
0.634
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr2_-_165406098
|
0.633
|
|
COBLL1
|
COBL-like 1
|
|
chr11_-_66898223
|
0.632
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr16_-_30445548
|
0.628
|
|
|
|
|
chr20_-_17610704
|
0.624
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr6_-_44341296
|
0.621
|
NM_004556
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr14_+_23907065
|
0.617
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr2_+_48521411
|
0.616
|
NM_001135629
NM_001193475
NM_152994
|
KLRAQ1
|
KLRAQ motif containing 1
|
|
chr7_+_73080362
|
0.616
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr6_+_44299254
|
0.615
|
NM_001078175
NM_001078176
NM_001078177
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr2_+_20729904
|
0.615
|
NM_182828
|
GDF7
|
growth differentiation factor 7
|
|
chr14_-_30565275
|
0.613
|
NM_001083893
NM_014574
|
STRN3
|
striatin, calmodulin binding protein 3
|
|
chr10_-_135021502
|
0.610
|
NM_001098483
NM_198472
|
C10orf125
|
chromosome 10 open reading frame 125
|
|
chr2_-_74634569
|
0.609
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr21_+_46473494
|
0.608
|
|
MCM3AP-AS1
|
MCM3AP antisense RNA 1 (non-protein coding)
|
|
chr14_+_23905984
|
0.607
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr12_+_732349
|
0.605
|
NM_001184985
NM_014823
NM_018979
NM_213655
|
WNK1
|
WNK lysine deficient protein kinase 1
|
|
chr9_+_33807213
|
0.604
|
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr9_-_112840801
|
0.604
|
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr19_-_11355882
|
0.602
|
NM_000121
|
EPOR
|
erythropoietin receptor
|
|
chr14_-_88953059
|
0.602
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr3_+_58198268
|
0.600
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr20_-_60375695
|
0.599
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr2_-_219960725
|
0.598
|
|
DNPEP
|
aspartyl aminopeptidase
|
|
chr14_-_68689058
|
0.597
|
|
DCAF5
|
DDB1 and CUL4 associated factor 5
|
|
chr4_-_74343320
|
0.596
|
NM_032217
NM_198889
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr3_+_58198455
|
0.591
|
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr10_+_94439658
|
0.591
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr16_+_2503978
|
0.590
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr17_+_26446067
|
0.589
|
NM_000267
NM_001042492
NM_001128147
|
NF1
|
neurofibromin 1
|
|
chr6_+_33486750
|
0.587
|
NM_002636
NM_024165
|
PHF1
|
PHD finger protein 1
|
|
chr5_+_122452739
|
0.587
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr2_-_165406173
|
0.586
|
NM_014900
|
COBLL1
|
COBL-like 1
|
|
chr5_-_175969697
|
0.576
|
NM_052899
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr19_+_54309423
|
0.576
|
NM_022165
|
LIN7B
|
lin-7 homolog B (C. elegans)
|
|
chr11_+_56984702
|
0.576
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr10_+_94823636
|
0.574
|
NM_000783
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr14_+_95575319
|
0.573
|
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr17_-_40401138
|
0.572
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr3_+_171238577
|
0.571
|
|
GPR160
|
G protein-coupled receptor 160
|
|
chr3_+_171238476
|
0.568
|
|
GPR160
|
G protein-coupled receptor 160
|
|
chr20_+_48240772
|
0.566
|
NM_005194
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr11_-_65082208
|
0.565
|
NM_001130144
NM_001164266
NM_021070
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr16_-_10183616
|
0.558
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr8_+_37672427
|
0.558
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr2_+_136005423
|
0.556
|
NM_015361
|
R3HDM1
|
R3H domain containing 1
|
|
chr12_-_61614930
|
0.553
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|