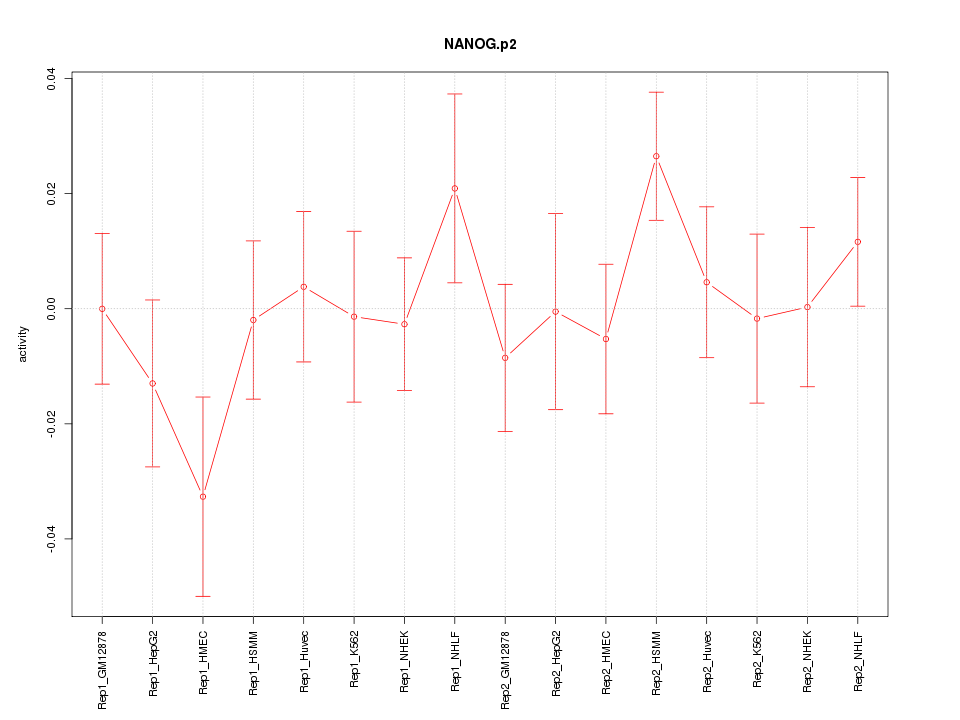
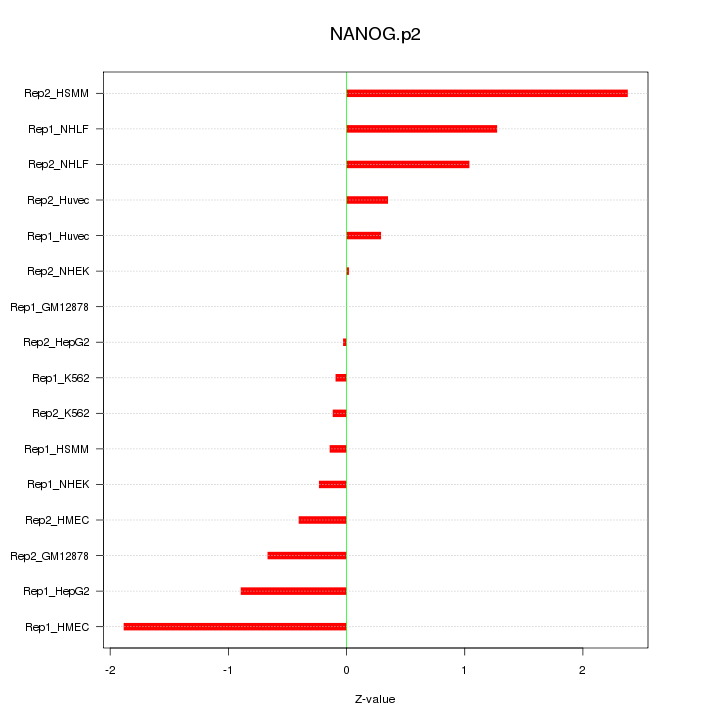
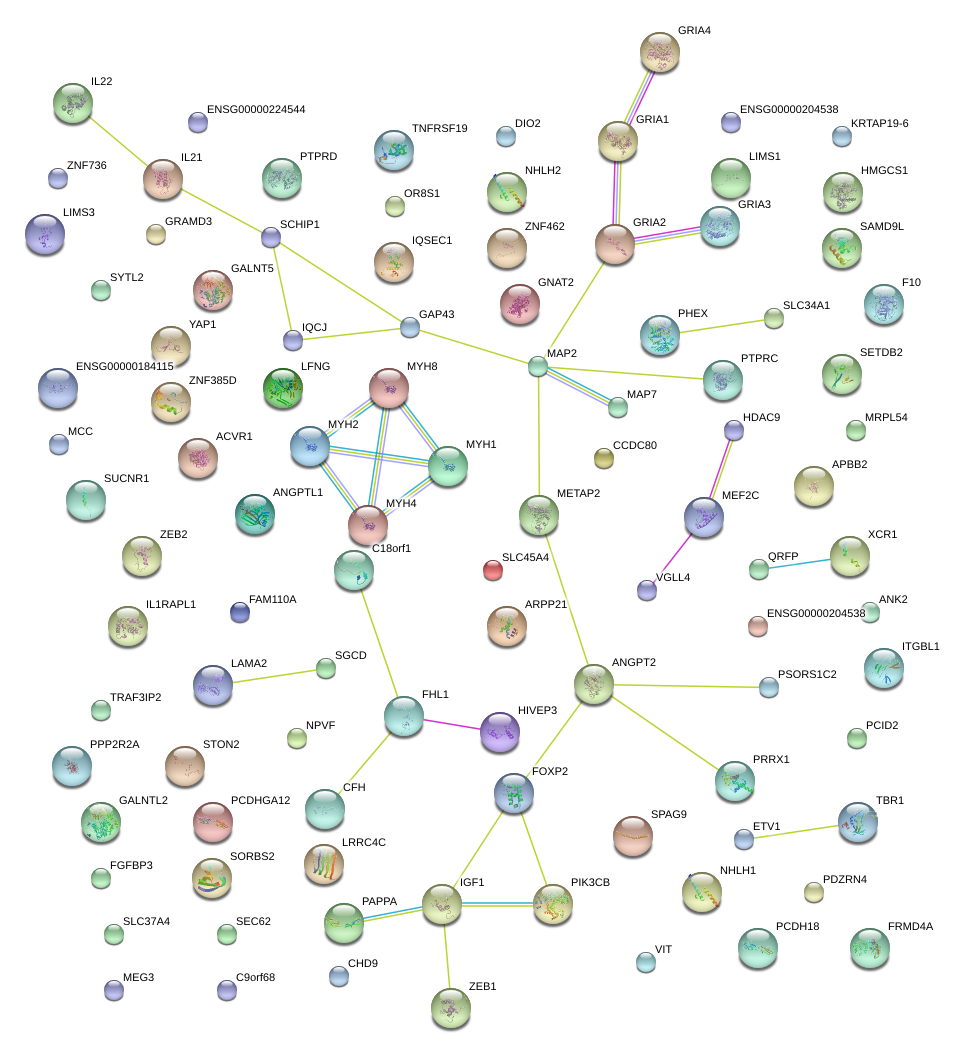
|
chr1_-_177106702
|
1.356
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr14_+_100365162
|
1.313
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chrX_+_28515479
|
1.265
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr7_-_13996166
|
1.205
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr2_-_144994038
|
1.077
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_110665188
|
1.043
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr4_+_114190233
|
0.928
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr9_+_117955890
|
0.916
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr5_+_176744037
|
0.889
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr4_-_186815116
|
0.884
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr9_-_8723893
|
0.881
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr1_+_168898898
|
0.865
|
|
PRRX1
|
paired related homeobox 1
|
|
chr16_+_51690564
|
0.839
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr21_-_30836051
|
0.823
|
NM_181612
|
KRTAP19-6
|
keratin associated protein 19-6
|
|
chr5_+_155686333
|
0.791
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr13_+_100902942
|
0.739
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr3_+_161040343
|
0.698
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_+_116824840
|
0.689
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr4_-_138673014
|
0.671
|
|
PCDH18
|
protocadherin 18
|
|
chr4_+_114433551
|
0.647
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr6_+_129245978
|
0.635
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr9_+_108665168
|
0.635
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr6_-_112034166
|
0.634
|
NM_001164281
NM_147686
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr4_-_40631397
|
0.619
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr5_+_152850276
|
0.592
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr2_+_210152399
|
0.572
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr3_-_11598829
|
0.562
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr13_+_112825113
|
0.557
|
NM_000504
|
F10
|
coagulation factor X
|
|
chr7_-_13992351
|
0.550
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr14_+_100365390
|
0.534
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr5_-_112658200
|
0.534
|
|
MCC
|
mutated in colorectal cancers
|
|
chrX_+_55951280
|
0.534
|
|
|
|
|
chr3_-_113842652
|
0.531
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr2_+_157822355
|
0.522
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr3_+_161040439
|
0.521
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr6_+_39868116
|
0.520
|
|
|
|
|
chr3_+_16191159
|
0.520
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr13_+_23042722
|
0.515
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr7_+_18501876
|
0.515
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr1_-_42156957
|
0.500
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr19_+_3713650
|
0.499
|
NM_172251
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr10_-_13789628
|
0.493
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr7_-_25234582
|
0.479
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr6_-_136889302
|
0.463
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr5_-_112658456
|
0.462
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr1_+_196874839
|
0.459
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr6_-_136888771
|
0.457
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr5_+_125786717
|
0.456
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr5_+_140790141
|
0.455
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr7_+_2524022
|
0.452
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr1_-_42157077
|
0.451
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr2_+_161980865
|
0.450
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr2_+_36777336
|
0.447
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr4_-_138673078
|
0.445
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr11_-_85107569
|
0.444
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr9_-_132759045
|
0.441
|
NM_198180
|
QRFP
|
pyroglutamylated RFamide peptide
|
|
chr1_+_559618
|
0.433
|
|
|
|
|
chr1_-_42156913
|
0.433
|
|
|
|
|
chr14_-_79748273
|
0.429
|
NM_000793
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr17_-_46479127
|
0.426
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr12_-_120725210
|
0.425
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr3_+_35697432
|
0.422
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_+_18502342
|
0.404
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr9_-_4656665
|
0.401
|
NM_001039395
|
C9orf68
|
chromosome 9 open reading frame 68
|
|
chr7_+_63411813
|
0.400
|
|
ZNF736
|
zinc finger protein 736
|
|
chr3_-_12916547
|
0.399
|
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr8_+_26206648
|
0.399
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr1_+_196874759
|
0.395
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr2_+_36777406
|
0.388
|
|
VIT
|
vitrin
|
|
chr2_+_210152609
|
0.386
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr2_+_108637916
|
0.373
|
|
LIMS1
LIMS3
|
LIM and senescent cell antigen-like domains 1
LIM and senescent cell antigen-like domains 3
|
|
chr3_-_21767819
|
0.371
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr1_-_109957195
|
0.367
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr2_-_158439868
|
0.367
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr4_-_123761615
|
0.362
|
NM_021803
|
IL21
|
interleukin 21
|
|
chrX_+_21960841
|
0.362
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr13_+_48923688
|
0.360
|
NM_001160308
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr10_-_93659164
|
0.358
|
NM_152429
|
FGFBP3
|
fibroblast growth factor binding protein 3
|
|
chr3_-_139960874
|
0.357
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr18_+_13601664
|
0.351
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr22_+_44828338
|
0.351
|
|
|
|
|
chr6_-_31215089
|
0.350
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr7_-_92615593
|
0.346
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr12_-_101398451
|
0.342
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_116185215
|
0.342
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr11_-_40272239
|
0.339
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr5_-_43326922
|
0.339
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr5_+_140790310
|
0.331
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr8_-_142333339
|
0.330
|
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr13_-_59223074
|
0.330
|
|
|
|
|
chr7_+_113842470
|
0.325
|
|
FOXP2
|
forkhead box P2
|
|
chr3_+_171183203
|
0.323
|
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr12_-_101398470
|
0.321
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr3_+_153074118
|
0.316
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr5_-_88215034
|
0.315
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chrX_+_135058402
|
0.312
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_-_118405288
|
0.310
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr12_-_101398429
|
0.309
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr12_+_40117751
|
0.309
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr12_-_101398503
|
0.308
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr8_-_6407882
|
0.306
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr12_+_47205681
|
0.305
|
NM_001005203
|
OR8S1
|
olfactory receptor, family 8, subfamily S, member 1
|
|
chr11_+_101488380
|
0.302
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr3_-_46043982
|
0.298
|
NM_001024644
NM_005283
|
XCR1
|
chemokine (C motif) receptor 1
|
|
chr17_-_10313600
|
0.298
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr14_-_80934523
|
0.297
|
NM_033104
|
STON2
|
stonin 2
|
|
chr2_+_161981046
|
0.296
|
|
TBR1
|
T-box, brain, 1
|
|
chr10_-_52315436
|
0.294
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr3_-_115960807
|
0.294
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_125786975
|
0.290
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr2_+_161981138
|
0.290
|
|
TBR1
|
T-box, brain, 1
|
|
chr12_-_4358968
|
0.287
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr17_+_56843893
|
0.284
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr5_-_96504216
|
0.282
|
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr3_+_130014957
|
0.279
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr5_+_140848905
|
0.276
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr5_+_140790365
|
0.274
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr1_-_20682541
|
0.273
|
|
|
|
|
chr6_-_32605898
|
0.272
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr10_+_11246934
|
0.270
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr11_+_7574992
|
0.269
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr12_-_101398288
|
0.268
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr6_-_114490701
|
0.267
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr11_-_104421240
|
0.267
|
NM_001017534
NM_052889
|
CARD16
CASP1
|
caspase recruitment domain family, member 16
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr4_+_71806538
|
0.263
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr5_-_96504194
|
0.263
|
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr2_-_224175386
|
0.263
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr15_-_68177283
|
0.262
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr6_-_112033775
|
0.262
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chrX_+_114701432
|
0.260
|
NM_005032
|
PLS3
|
plastin 3
|
|
chr17_-_45629401
|
0.259
|
|
|
|
|
chr22_-_37211467
|
0.257
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr3_-_69254226
|
0.254
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr7_+_28692122
|
0.254
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_+_54700955
|
0.253
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr11_-_85108030
|
0.252
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr11_-_66976775
|
0.251
|
NM_206997
|
GPR152
|
G protein-coupled receptor 152
|
|
chr7_-_13995811
|
0.249
|
|
ETV1
|
ets variant 1
|
|
chr18_+_71051713
|
0.248
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr18_+_54039779
|
0.248
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_-_45621571
|
0.247
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr7_+_116447527
|
0.246
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr8_-_82557956
|
0.243
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr3_+_2528272
|
0.240
|
|
CNTN4
|
contactin 4
|
|
chr8_+_26569709
|
0.240
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_121445831
|
0.239
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr12_-_69434330
|
0.239
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr5_+_53787187
|
0.237
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr3_-_113842553
|
0.237
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr12_+_54838408
|
0.236
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr2_-_224175339
|
0.235
|
|
SCG2
|
secretogranin II
|
|
chr12_-_89922888
|
0.234
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr7_+_113853790
|
0.231
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr3_-_55489973
|
0.229
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr11_-_128217550
|
0.227
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr18_+_54039818
|
0.227
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr14_+_99309688
|
0.227
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr4_+_169250281
|
0.224
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chrX_+_102918409
|
0.224
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr15_-_91078307
|
0.223
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr9_+_81377289
|
0.222
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr1_+_50347188
|
0.221
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr17_-_36418869
|
0.221
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr12_+_25096755
|
0.221
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr18_+_30812205
|
0.218
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr11_-_85074848
|
0.218
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chrX_+_134482387
|
0.217
|
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr12_-_14740694
|
0.216
|
NM_004963
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor)
|
|
chr5_-_96504258
|
0.216
|
NM_153234
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr4_+_15038726
|
0.216
|
NM_031911
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr1_-_149004916
|
0.215
|
NM_004079
|
CTSS
|
cathepsin S
|
|
chrX_+_41433169
|
0.215
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr17_-_45621312
|
0.214
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_-_149004852
|
0.214
|
|
CTSS
|
cathepsin S
|
|
chr17_+_51585834
|
0.213
|
NM_153228
|
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1
|
|
chr7_-_136679045
|
0.212
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr2_+_108637698
|
0.212
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr9_+_123369219
|
0.209
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr1_-_203579932
|
0.208
|
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr2_-_224175293
|
0.208
|
|
SCG2
|
secretogranin II
|
|
chr1_+_194887630
|
0.206
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr2_-_200033445
|
0.205
|
|
SATB2
|
SATB homeobox 2
|
|
chr14_-_104330982
|
0.204
|
NM_005163
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr5_-_148738980
|
0.204
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr18_-_3864252
|
0.203
|
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr3_-_102194938
|
0.203
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr3_+_157877146
|
0.201
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr9_-_4289574
|
0.200
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr12_+_54838733
|
0.200
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr5_-_24535386
|
0.195
|
NM_001190450
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr15_+_100175888
|
0.195
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chr20_+_42463279
|
0.194
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr2_-_217268492
|
0.194
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr19_-_38052481
|
0.193
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr1_-_115060956
|
0.193
|
NM_002524
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr5_-_168204430
|
0.192
|
|
|
|
|
chr1_-_115060857
|
0.192
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr17_-_37590910
|
0.191
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr7_+_113842284
|
0.191
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr12_-_89922875
|
0.191
|
|
EPYC
|
epiphycan
|