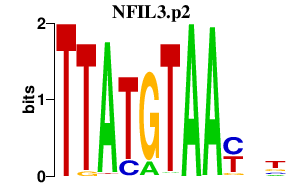
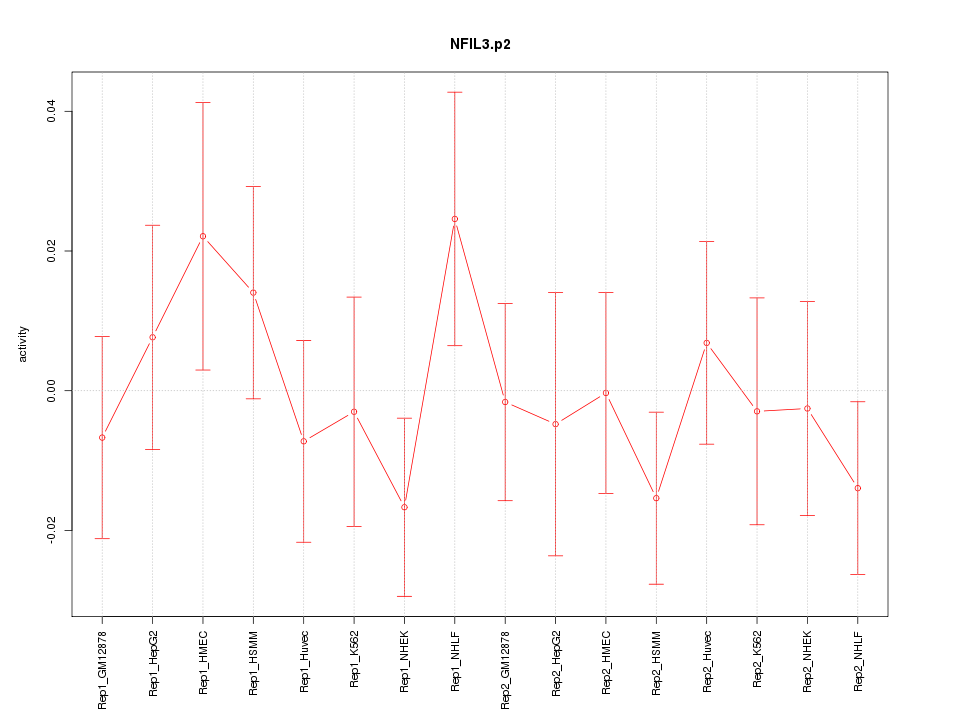
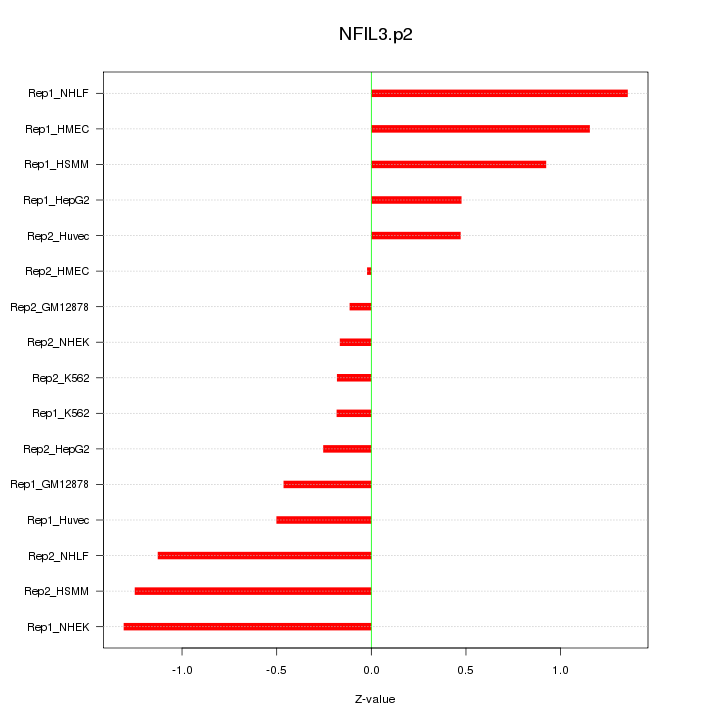
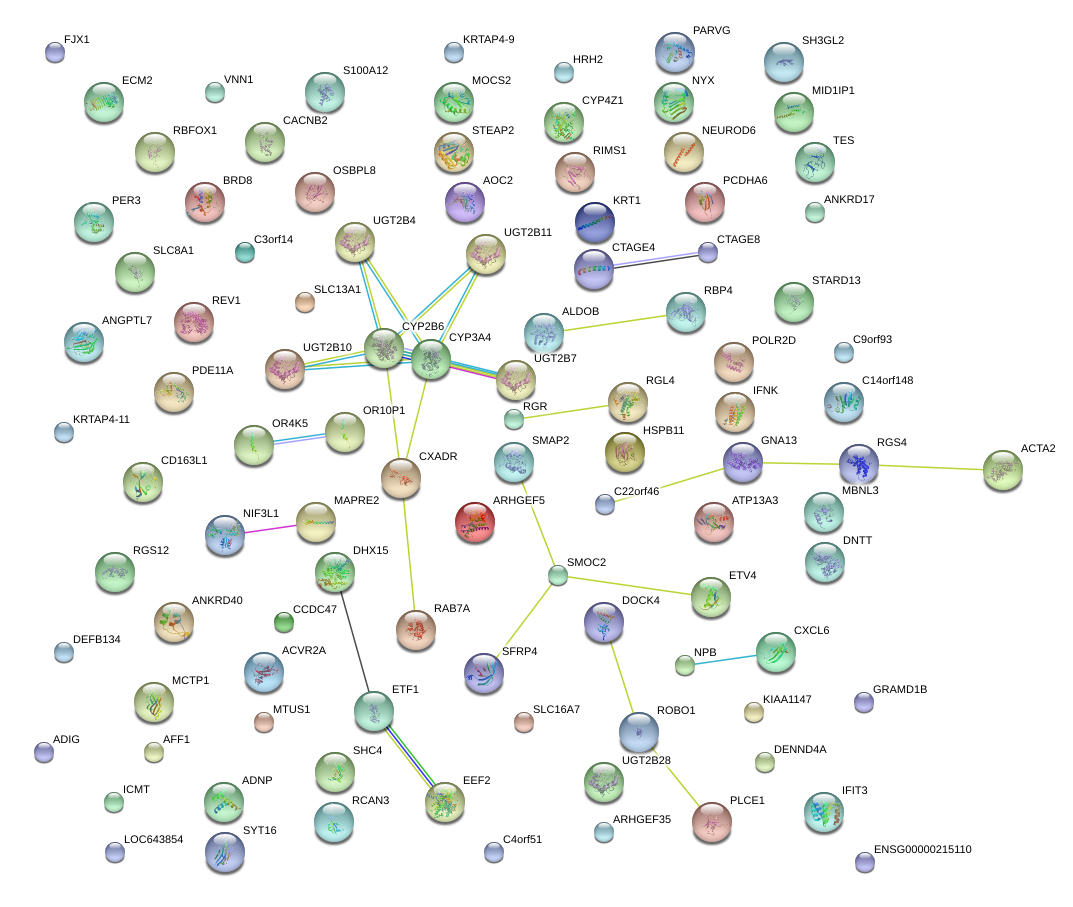
|
chr19_-_3936460
|
0.992
|
NM_001961
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr10_+_95838801
|
0.924
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr6_+_72979234
|
0.909
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr22_+_42900168
|
0.788
|
NM_001137605
|
PARVG
|
parvin, gamma
|
|
chr17_-_46140015
|
0.783
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr9_-_94338072
|
0.720
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr22_-_34564611
|
0.718
|
|
RPL41P5
|
ribosomal protein L41 pseudogene 5
|
|
chr9_+_15542871
|
0.700
|
NM_173550
|
C9orf93
|
chromosome 9 open reading frame 93
|
|
chr1_+_24713396
|
0.690
|
|
RCAN3
|
RCAN family member 3
|
|
chr17_+_38250122
|
0.645
|
NM_001158
NM_009590
|
AOC2
|
amine oxidase, copper containing 2 (retina-specific)
|
|
chr10_+_18589589
|
0.582
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr7_-_31347032
|
0.581
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr14_+_61532293
|
0.574
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr9_+_27514311
|
0.550
|
NM_020124
|
IFNK
|
interferon, kappa
|
|
chr14_-_76958984
|
0.545
|
NM_138791
NM_001113475
|
C14orf148
|
chromosome 14 open reading frame 148
|
|
chr4_-_74307637
|
0.527
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr13_-_32678142
|
0.514
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_+_54316910
|
0.492
|
NM_206899
|
OR10P1
|
olfactory receptor, family 10, subfamily P, member 1
|
|
chr1_+_11171967
|
0.482
|
NM_021146
|
ANGPTL7
|
angiopoietin-like 7
|
|
chr1_+_47305633
|
0.482
|
NM_178134
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1
|
|
chr20_-_48980858
|
0.475
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr5_-_137876416
|
0.469
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr5_+_140227951
|
0.467
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chrX_+_41191630
|
0.458
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr7_+_89681069
|
0.451
|
NM_001040666
|
STEAP2
|
six transmembrane epithelial antigen of the prostate 2
|
|
chr4_+_47403107
|
0.450
|
|
|
|
|
chr6_-_133076873
|
0.442
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr2_-_178461710
|
0.439
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr4_+_146820805
|
0.427
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr8_-_11891168
|
0.423
|
NM_001033019
|
DEFB134
|
defensin, beta 134
|
|
chr4_+_88187215
|
0.420
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr11_+_35596310
|
0.416
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chr7_-_111216180
|
0.416
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr11_+_122936246
|
0.396
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr5_-_94443300
|
0.389
|
NM_001002796
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr10_-_95350949
|
0.382
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr9_+_17568952
|
0.380
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr15_-_47042932
|
0.379
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr1_-_6218576
|
0.374
|
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr10_+_91077581
|
0.369
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr17_+_36515166
|
0.349
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr11_-_10487212
|
0.347
|
NM_001190702
|
MTRNR2L8
|
MT-RNR2-like 8
|
|
chr11_+_122936145
|
0.347
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr17_-_59204622
|
0.347
|
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr21_+_17841297
|
0.343
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr13_+_96797290
|
0.342
|
|
|
|
|
chr5_-_52441044
|
0.332
|
NM_004531
|
MOCS2
|
molybdenum cofactor synthesis 2
|
|
chr4_+_3341521
|
0.332
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr2_-_99472843
|
0.329
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr17_-_38979178
|
0.326
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr3_-_195670256
|
0.322
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr7_-_122627235
|
0.304
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr1_-_6218554
|
0.300
|
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr14_+_19458580
|
0.297
|
NM_001005483
|
OR4K5
|
olfactory receptor, family 4, subfamily K, member 5
|
|
chr17_-_59204685
|
0.296
|
NM_020198
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr2_+_201464884
|
0.293
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. pombe)
|
|
chr7_-_143597313
|
0.288
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr3_-_78749633
|
0.285
|
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr18_+_30875297
|
0.285
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr19_+_46189022
|
0.281
|
NM_000767
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr2_+_148319033
|
0.281
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr12_-_51360439
|
0.278
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr3_+_130014957
|
0.275
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr8_-_17599438
|
0.272
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr4_+_74921136
|
0.270
|
NM_002993
|
CXCL6
|
chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2)
|
|
chr7_-_141048358
|
0.266
|
NM_001080392
|
KIAA1147
|
KIAA1147
|
|
chr17_-_36528113
|
0.259
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr20_+_36643251
|
0.259
|
NM_001018082
|
ADIG
|
adipogenin
|
|
chr12_-_7487989
|
0.259
|
NM_174941
|
CD163L1
|
CD163 molecule-like 1
|
|
chr17_-_60439764
|
0.248
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr12_-_123568781
|
0.248
|
|
|
|
|
chr15_-_63871492
|
0.245
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr16_+_7322751
|
0.243
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr2_-_40510923
|
0.242
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chrX_-_131375217
|
0.238
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr10_+_85994788
|
0.237
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr4_-_70115037
|
0.235
|
NM_001073
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11
|
|
chr2_-_99472910
|
0.234
|
NM_001037872
NM_016316
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr5_-_79982584
|
0.233
|
NM_001190470
|
MTRNR2L2
|
MT-RNR2-like 2
|
|
chr5_+_175041069
|
0.229
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr1_-_151614681
|
0.225
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chrX_+_38548016
|
0.225
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr7_+_115650240
|
0.224
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr10_+_98054074
|
0.224
|
NM_001017520
NM_004088
|
DNTT
|
deoxynucleotidyltransferase, terminal
|
|
chr3_+_62280435
|
0.223
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr9_-_103237851
|
0.223
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr1_-_54183833
|
0.222
|
NM_016126
|
HSPB11
|
heat shock protein family B (small), member 11
|
|
chr1_+_7766966
|
0.219
|
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr1_+_40611955
|
0.216
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr7_-_37923040
|
0.214
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr12_+_58369392
|
0.214
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr1_+_161305188
|
0.211
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr22_+_40416492
|
0.211
|
NM_001142964
|
C22orf46
|
chromosome 22 open reading frame 46
|
|
chr11_+_6823458
|
0.206
|
NM_178168
|
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5
|
|
chr1_+_40612016
|
0.204
|
|
SMAP2
|
small ArfGAP2
|
|
chr5_-_147996769
|
0.198
|
NM_001040169
NM_001040172
NM_001040173
NM_199453
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr17_+_36493927
|
0.195
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr13_+_96672541
|
0.193
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_+_93394025
|
0.191
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr17_+_55269974
|
0.190
|
|
|
|
|
chr11_-_83661878
|
0.189
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr10_-_14412813
|
0.187
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr17_+_55098280
|
0.185
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr17_-_34209671
|
0.183
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr5_+_86712376
|
0.182
|
|
|
|
|
chr4_-_96689373
|
0.181
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr6_+_154449334
|
0.181
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr2_-_202811459
|
0.181
|
NM_001005781
NM_001005782
NM_003352
|
SUMO1
|
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae)
|
|
chr1_-_177106702
|
0.178
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr5_-_94442941
|
0.172
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr6_-_26151863
|
0.172
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr1_+_57093030
|
0.170
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr12_-_90029328
|
0.169
|
|
LUM
|
lumican
|
|
chr6_+_7347878
|
0.167
|
NM_153005
|
RIOK1
|
RIO kinase 1 (yeast)
|
|
chr2_-_86276102
|
0.166
|
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr13_-_37070862
|
0.165
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr6_-_132073849
|
0.164
|
NM_001145659
|
CTAGE9
|
CTAGE family, member 9
|
|
chr1_+_194887630
|
0.164
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr19_+_46189124
|
0.163
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr2_-_127817143
|
0.163
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr19_+_46286207
|
0.162
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr21_-_30796278
|
0.162
|
NM_181611
|
KRTAP19-5
|
keratin associated protein 19-5
|
|
chr1_-_42157077
|
0.159
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr1_+_86785346
|
0.159
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr6_+_158654890
|
0.159
|
|
|
|
|
chr3_+_73193499
|
0.153
|
NM_018029
|
EBLN2
|
endogenous Borna-like N element-2
|
|
chr5_+_172415891
|
0.153
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr9_+_71009746
|
0.152
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr19_-_58486686
|
0.152
|
NM_033341
|
BIRC8
|
baculoviral IAP repeat containing 8
|
|
chrX_-_77037563
|
0.152
|
NM_032121
|
MAGT1
|
magnesium transporter 1
|
|
chr16_-_54424517
|
0.151
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr9_+_96561750
|
0.150
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr2_-_188021117
|
0.150
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr10_-_64698831
|
0.149
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr3_-_192063158
|
0.145
|
NM_001146686
|
GEMC1
|
geminin coiled-coil domain-containing protein 1
|
|
chr1_+_202033273
|
0.144
|
NM_001174108
|
ZBED6
|
zinc finger, BED-type containing 6
|
|
chr3_+_171167243
|
0.142
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr8_-_91066060
|
0.141
|
NM_002485
|
NBN
|
nibrin
|
|
chr11_-_62539857
|
0.141
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chr4_-_69218909
|
0.139
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr15_+_91244422
|
0.138
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr10_-_82039158
|
0.137
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr15_+_91244563
|
0.137
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr6_-_123999640
|
0.134
|
NM_006073
|
TRDN
|
triadin
|
|
chr18_+_19826734
|
0.134
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr7_+_134114701
|
0.134
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr4_-_186815116
|
0.134
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_167445284
|
0.133
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr22_-_48607199
|
0.133
|
|
BRD1
|
bromodomain containing 1
|
|
chr8_-_72437020
|
0.133
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr22_+_25347927
|
0.133
|
NM_001886
|
CRYBA4
|
crystallin, beta A4
|
|
chr1_+_144152864
|
0.129
|
|
TXNIP
|
thioredoxin interacting protein
|
|
chr9_+_83748234
|
0.129
|
NM_207416
|
FAM75D3
|
family with sequence similarity 75, member D3
|
|
chr12_+_15955499
|
0.129
|
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr11_+_64551472
|
0.127
|
NM_013306
NM_147777
|
SNX15
|
sorting nexin 15
|
|
chr17_-_1195096
|
0.127
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr4_+_77214886
|
0.127
|
NM_001130016
NM_001179
|
ART3
|
ADP-ribosyltransferase 3
|
|
chr1_+_227506751
|
0.126
|
NM_006542
|
SPHAR
|
S-phase response (cyclin related)
|
|
chr8_-_130868315
|
0.126
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr2_-_192420168
|
0.125
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr2_-_224611568
|
0.123
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr8_+_67751007
|
0.123
|
NM_019607
|
C8orf44
|
chromosome 8 open reading frame 44
|
|
chr1_+_177261627
|
0.122
|
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr3_-_102888090
|
0.121
|
NM_000986
|
RPL24
|
ribosomal protein L24
|
|
chr21_-_14840506
|
0.117
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr1_+_148089251
|
0.117
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr9_+_32773496
|
0.116
|
NM_212558
|
TMEM215
|
transmembrane protein 215
|
|
chr10_+_90474280
|
0.116
|
NM_001080518
|
LIPK
|
lipase, family member K
|
|
chr8_-_102286695
|
0.116
|
|
ZNF706
|
zinc finger protein 706
|
|
chr1_+_177261678
|
0.116
|
NM_014864
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr9_-_32540818
|
0.114
|
|
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase
|
|
chr12_-_73889777
|
0.112
|
NM_139136
NM_139137
NM_153748
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr1_+_210805319
|
0.111
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr2_+_161701711
|
0.111
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr7_+_143511480
|
0.110
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr5_+_140235114
|
0.109
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr6_-_66346417
|
0.109
|
NM_198283
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr6_-_132987106
|
0.109
|
NM_001033080
|
TAAR2
|
trace amine associated receptor 2
|
|
chr1_+_151081953
|
0.109
|
NM_001128600
|
LCE6A
|
late cornified envelope 6A
|
|
chr7_+_143646150
|
0.107
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr3_+_103029523
|
0.107
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr2_+_169637255
|
0.106
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr12_-_44407515
|
0.106
|
|
LOC400027
|
hypothetical LOC400027
|
|
chr2_-_86276015
|
0.105
|
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr6_+_64289585
|
0.105
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr1_+_176577228
|
0.105
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr8_+_67568044
|
0.105
|
NM_152765
|
C8orf46
|
chromosome 8 open reading frame 46
|
|
chr12_+_54106304
|
0.103
|
NM_001005183
|
OR6C76
|
olfactory receptor, family 6, subfamily C, member 76
|
|
chr17_-_23965305
|
0.102
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr7_+_142723340
|
0.102
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr2_-_45648608
|
0.102
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr1_-_154988045
|
0.101
|
|
HDGF
|
hepatoma-derived growth factor
|
|
chr11_+_65026382
|
0.099
|
|
|
|
|
chr4_-_157006827
|
0.099
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chrX_+_79562601
|
0.098
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr3_+_188568861
|
0.098
|
NM_022147
|
RTP4
|
receptor (chemosensory) transporter protein 4
|
|
chr16_+_74868676
|
0.098
|
NM_033401
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chr8_-_103735194
|
0.097
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chrX_+_56772391
|
0.097
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr4_+_70180805
|
0.094
|
NM_053039
|
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28
|