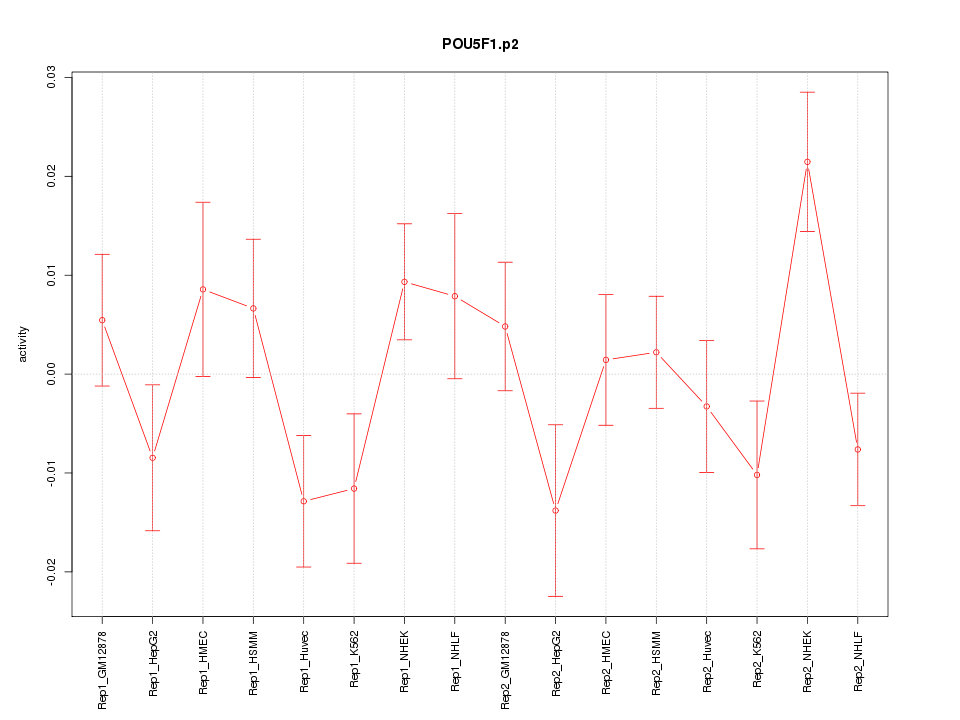
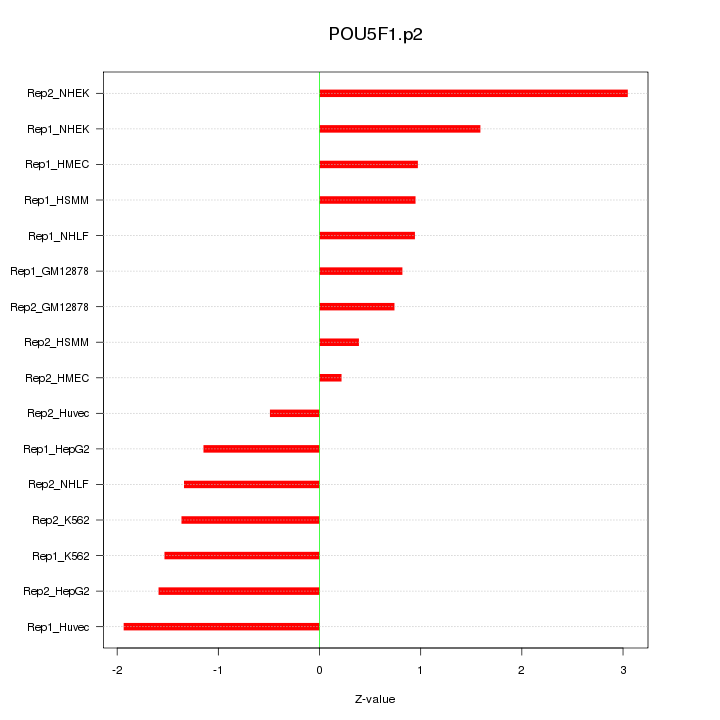
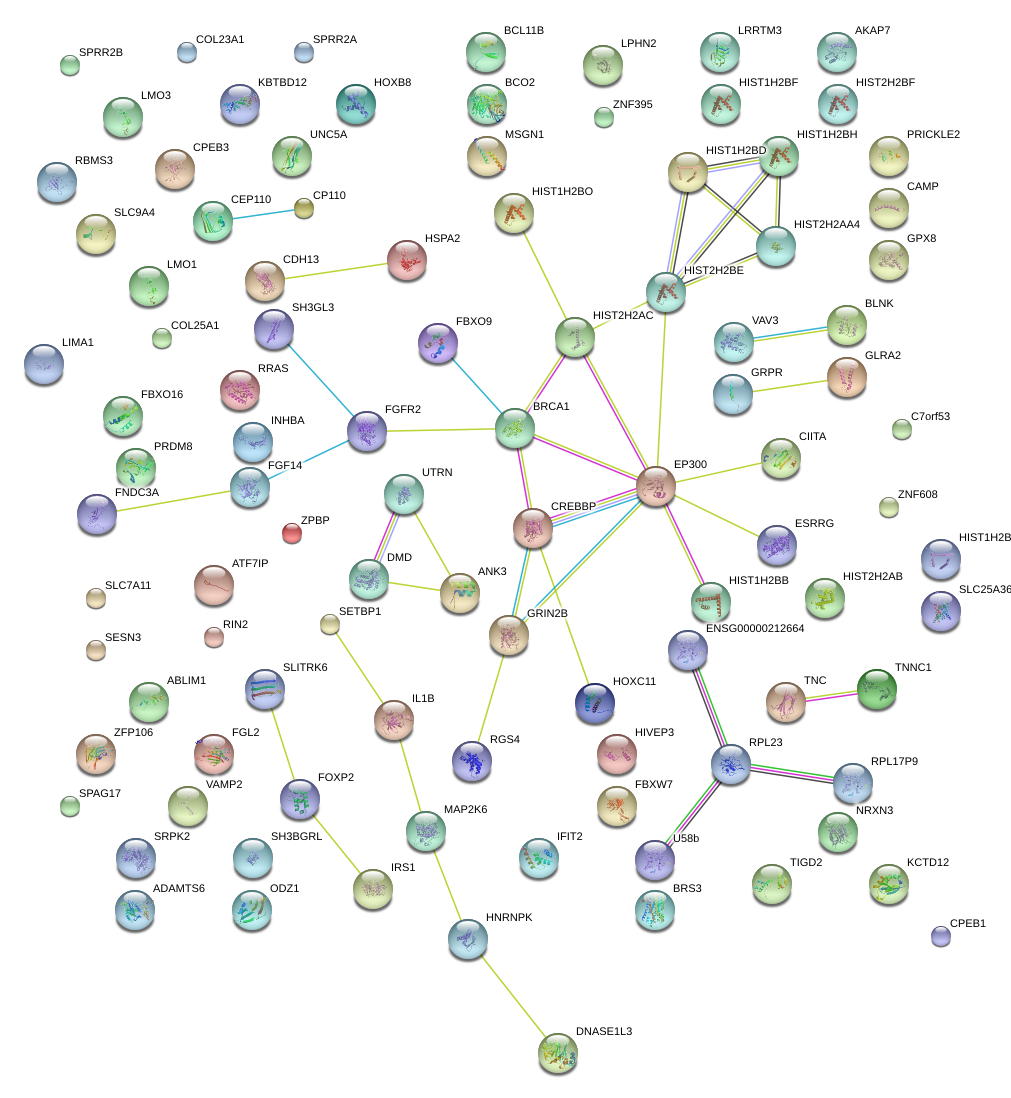
|
chr17_+_64922420
|
2.735
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr12_+_14409875
|
2.588
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chrX_-_33267646
|
2.527
|
NM_000109
|
DMD
|
dystrophin
|
|
chr3_-_64186151
|
2.463
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr13_-_101852028
|
1.965
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr10_+_68355797
|
1.753
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr7_-_76667045
|
1.668
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr5_+_54491702
|
1.665
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr13_-_85271329
|
1.644
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr1_+_161305188
|
1.470
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_-_48902692
|
1.432
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr1_+_161305558
|
1.410
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr16_-_3783475
|
1.391
|
|
CREBBP
|
CREB binding protein
|
|
chr18_-_45271938
|
1.383
|
|
RPL17-C18ORF32
RPL17
|
RPL17-C18orf32 read-through transcript
ribosomal protein L17
|
|
chrX_+_135397790
|
1.352
|
NM_001727
|
BRS3
|
bombesin-like receptor 3
|
|
chr19_-_54835162
|
1.311
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr6_-_26151863
|
1.304
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr1_-_215329569
|
1.291
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_+_81544432
|
1.169
|
|
LPHN2
|
latrophilin 2
|
|
chr3_-_58175437
|
1.158
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr11_+_111551417
|
1.116
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr3_+_142175329
|
1.092
|
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr2_+_102456193
|
1.062
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chrX_+_14457340
|
1.041
|
NM_001118885
NM_001118886
NM_001171942
NM_002063
|
GLRA2
|
glycine receptor, alpha 2
|
|
chr17_-_8004671
|
1.022
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr4_-_139382672
|
0.995
|
NM_014331
|
SLC7A11
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 11
|
|
chr9_-_116920232
|
0.949
|
NM_002160
|
TNC
|
tenascin C
|
|
chr12_+_52653176
|
0.947
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr10_-_98021144
|
0.941
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr14_+_64077171
|
0.934
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr20_+_19863716
|
0.907
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_+_161305019
|
0.907
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr5_-_64813459
|
0.894
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr10_-_62002672
|
0.885
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr3_+_29297806
|
0.878
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr3_+_129124591
|
0.874
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr1_-_151296611
|
0.865
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr7_-_50103335
|
0.865
|
NM_001159878
NM_007009
|
ZPBP
|
zona pellucida binding protein
|
|
chr7_-_104696678
|
0.863
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr15_-_81037554
|
0.857
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr4_+_81325447
|
0.855
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr12_-_14024187
|
0.852
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr6_+_131613196
|
0.850
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr5_-_177950161
|
0.842
|
NM_173465
|
COL23A1
|
collagen, type XXIII, alpha 1
|
|
chr7_+_113853790
|
0.841
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr15_-_40537002
|
0.834
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr7_+_111908143
|
0.834
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr18_+_40514772
|
0.832
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr5_-_124108672
|
0.813
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr13_-_76358370
|
0.811
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr4_-_153493133
|
0.795
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_-_118529303
|
0.789
|
NM_206996
|
SPAG17
|
sperm associated antigen 17
|
|
chr18_+_40513889
|
0.787
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr16_+_10878539
|
0.782
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr4_+_90252990
|
0.779
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr9_-_85773899
|
0.770
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr1_-_148126089
|
0.766
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr1_-_42156957
|
0.758
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr10_-_93992982
|
0.757
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr17_-_44047299
|
0.747
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr4_-_110443247
|
0.736
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr12_-_16649211
|
0.732
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr14_+_78815406
|
0.732
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr14_-_98807574
|
0.711
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr1_-_148050534
|
0.704
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chrX_+_16051344
|
0.704
|
NM_005314
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr2_+_17861266
|
0.700
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr13_+_48582389
|
0.688
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr7_-_41709191
|
0.686
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr11_+_65027721
|
0.683
|
|
|
|
|
chr1_-_42157077
|
0.681
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chrX_-_31436335
|
0.677
|
NM_004014
|
DMD
|
dystrophin
|
|
chr6_+_53043729
|
0.674
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr17_-_38529638
|
0.672
|
NM_007298
|
BRCA1
|
breast cancer 1, early onset
|
|
chrX_+_80344253
|
0.672
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr2_-_227371698
|
0.667
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr1_-_197173159
|
0.664
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr8_-_28403652
|
0.661
|
NM_172366
|
FBXO16
ZNF395
|
F-box protein 16
zinc finger protein 395
|
|
chr15_+_81907176
|
0.659
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr3_+_48239865
|
0.653
|
NM_004345
|
CAMP
|
cathelicidin antimicrobial peptide
|
|
chr16_+_81218142
|
0.651
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr10_-_123346148
|
0.646
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_-_113310796
|
0.640
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr15_+_81906983
|
0.635
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr11_-_94603857
|
0.632
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr9_+_122923864
|
0.631
|
|
CEP110
|
centrosomal protein 110kDa
|
|
chr10_+_91051685
|
0.630
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chrX_-_123925343
|
0.628
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1(Drosophila)
|
|
chr1_-_108308997
|
0.625
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr10_-_116276651
|
0.621
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr18_+_75968133
|
0.620
|
|
ADNP2
|
ADNP homeobox 2
|
|
chrX_-_62891066
|
0.611
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr2_+_237143118
|
0.611
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr2_+_173794692
|
0.611
|
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr5_+_49998528
|
0.609
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr11_+_69176391
|
0.599
|
|
CCND1
|
cyclin D1
|
|
chr2_-_218739891
|
0.597
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr11_-_34336102
|
0.595
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr5_+_49998677
|
0.588
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr14_-_93512818
|
0.585
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr4_+_41395493
|
0.585
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_73973004
|
0.585
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr22_-_44828636
|
0.584
|
NM_018280
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr4_+_120276386
|
0.581
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr3_+_51837608
|
0.578
|
NM_001085479
|
IQCF3
|
IQ motif containing F3
|
|
chr10_+_95838801
|
0.575
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr8_+_11703368
|
0.575
|
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr21_-_35174742
|
0.573
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_+_42107711
|
0.571
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr3_-_102194938
|
0.567
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr13_-_59223074
|
0.566
|
|
|
|
|
chr22_-_26527469
|
0.566
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr16_+_81218074
|
0.558
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr13_-_98428244
|
0.557
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr7_-_83662152
|
0.552
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr11_-_114880275
|
0.548
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chrX_+_80344215
|
0.546
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr4_+_76700281
|
0.533
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chr17_-_2250967
|
0.524
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr1_-_205012348
|
0.523
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr2_+_5750229
|
0.521
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr6_-_119512035
|
0.520
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr1_-_151280201
|
0.519
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr11_+_10434056
|
0.518
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr7_+_93862137
|
0.516
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr4_-_159313613
|
0.511
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr2_-_165768798
|
0.508
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr12_+_111829121
|
0.507
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr4_-_41579325
|
0.506
|
|
|
|
|
chr2_-_210726539
|
0.504
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr8_-_72437020
|
0.504
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr6_-_10523424
|
0.503
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr1_+_208008517
|
0.501
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr7_-_27149750
|
0.500
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr17_-_78391178
|
0.498
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr14_-_74669864
|
0.497
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr17_-_45427565
|
0.493
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr6_-_134540666
|
0.492
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr18_+_30544193
|
0.491
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr6_-_130224108
|
0.489
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr2_+_165804157
|
0.489
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr7_+_28418668
|
0.488
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_-_25958556
|
0.488
|
|
EBF2
|
early B-cell factor 2
|
|
chrX_-_106846924
|
0.486
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_-_147665783
|
0.486
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr4_+_124540114
|
0.483
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr3_-_129023850
|
0.476
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr22_+_36422940
|
0.476
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr4_-_159313167
|
0.474
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr9_+_35722490
|
0.471
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr3_-_87122958
|
0.469
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr3_+_145174856
|
0.469
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr1_+_27541086
|
0.469
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr21_-_31123797
|
0.466
|
NM_181606
|
KRTAP7-1
|
keratin associated protein 7-1 (gene/pseudogene)
|
|
chr2_+_65517316
|
0.466
|
|
FLJ16124
|
FLJ16124 protein
|
|
chr5_-_172976212
|
0.465
|
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chrX_-_80344096
|
0.465
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr1_+_172683834
|
0.461
|
NM_005684
|
GPR52
|
G protein-coupled receptor 52
|
|
chrX_-_24575375
|
0.456
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr12_-_11066436
|
0.456
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr11_+_34599227
|
0.452
|
|
EHF
|
ets homologous factor
|
|
chr13_-_32757835
|
0.451
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_+_52713098
|
0.451
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr1_-_211087570
|
0.450
|
NM_001024601
|
C1orf227
|
chromosome 1 open reading frame 227
|
|
chr5_-_172976196
|
0.450
|
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chr6_-_10523203
|
0.449
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr18_-_51406841
|
0.449
|
NM_001083962
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr4_-_159312973
|
0.448
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_+_15145001
|
0.447
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr11_-_111539881
|
0.447
|
NM_001562
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor)
|
|
chr8_-_119192985
|
0.446
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr8_-_15140162
|
0.444
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr2_-_40593074
|
0.442
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr1_+_168381811
|
0.442
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr12_-_13139848
|
0.441
|
NM_031289
NM_153823
|
GSG1
|
germ cell associated 1
|
|
chr11_-_119513429
|
0.440
|
|
TRIM29
|
tripartite motif containing 29
|
|
chrX_-_106846684
|
0.440
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr10_-_64698831
|
0.436
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr5_-_172976259
|
0.433
|
NM_001159651
NM_138369
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chr6_+_26381122
|
0.431
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr6_+_35812836
|
0.430
|
NM_145028
|
C6orf81
|
chromosome 6 open reading frame 81
|
|
chr6_-_32892705
|
0.429
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chrX_+_28515479
|
0.428
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr2_+_143351536
|
0.424
|
NM_001032998
NM_003937
|
KYNU
|
kynureninase
|
|
chr4_-_100575313
|
0.423
|
NM_001166504
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr11_+_34599163
|
0.423
|
NM_012153
|
EHF
|
ets homologous factor
|
|
chr15_+_62578671
|
0.422
|
NM_015042
|
ZNF609
|
zinc finger protein 609
|
|
chr9_+_108665168
|
0.419
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr14_+_64076916
|
0.418
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr12_-_49897743
|
0.414
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr3_-_9266251
|
0.414
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr4_-_47534815
|
0.411
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr1_-_151310707
|
0.410
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr17_+_38306339
|
0.410
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chrX_-_109570113
|
0.408
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr6_-_42527760
|
0.408
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr1_+_200246312
|
0.406
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr8_+_123863169
|
0.406
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr8_-_93181091
|
0.406
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_+_104986009
|
0.406
|
NM_000829
NM_001077243
NM_001077244
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|