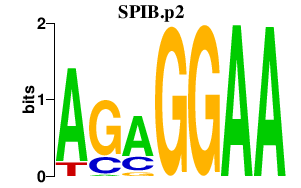
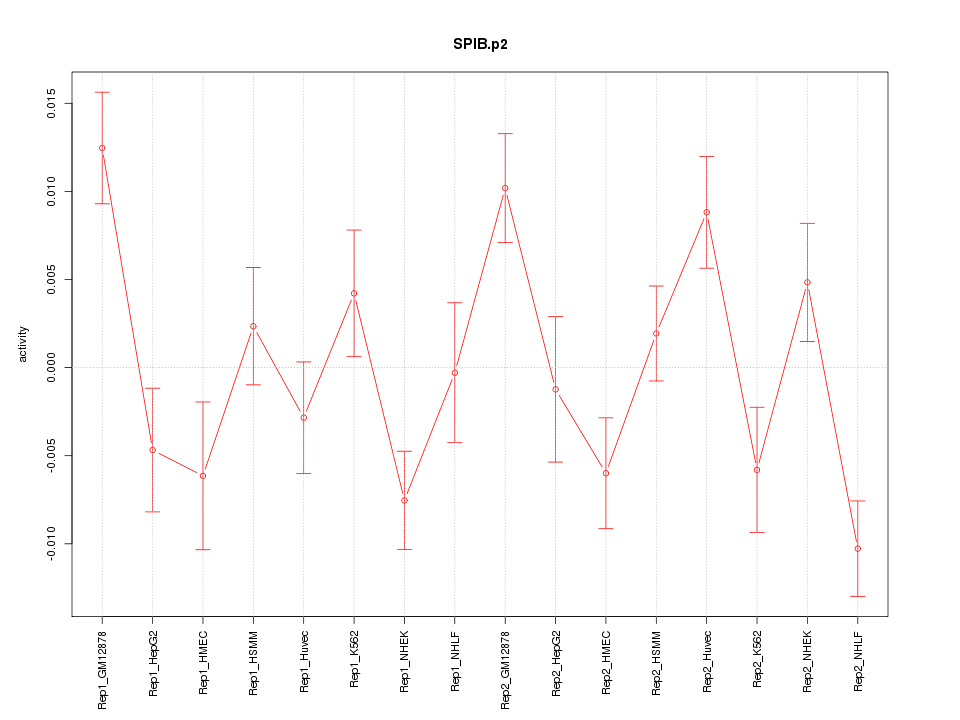
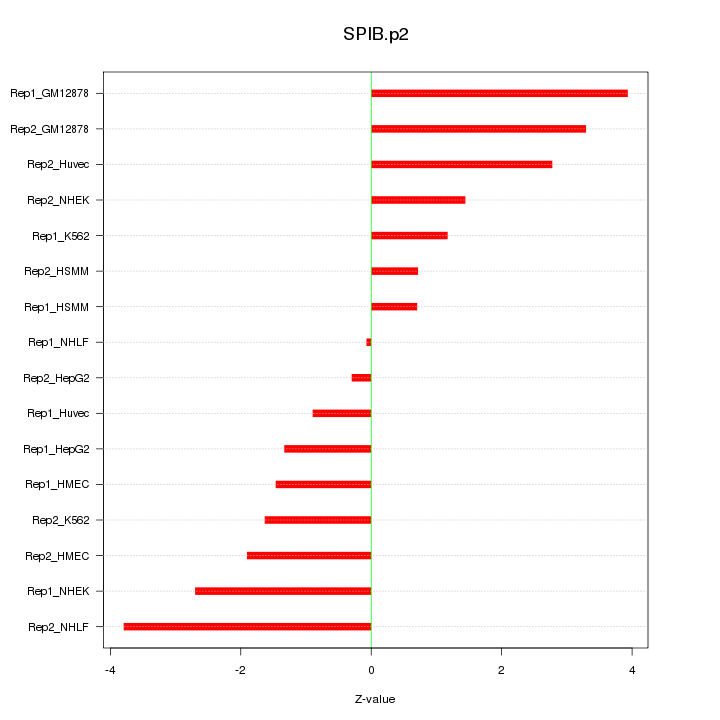
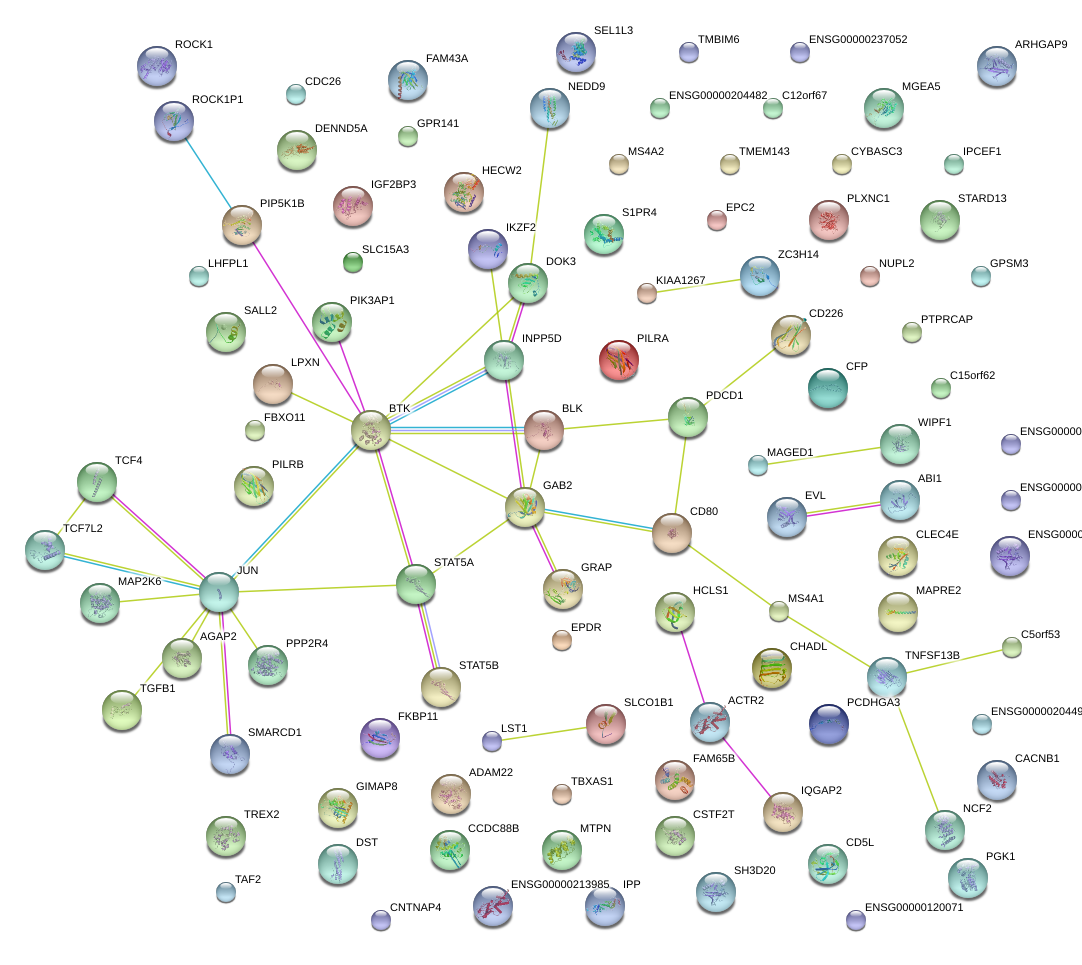
|
chr2_-_175207521
|
1.555
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr10_-_98470164
|
1.275
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr17_-_40843530
|
1.124
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr11_-_60880799
|
1.099
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr1_-_59020875
|
1.041
|
|
JUN
|
jun proto-oncogene
|
|
chr5_+_140772691
|
1.001
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr6_+_31661934
|
0.952
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr22_-_39964001
|
0.909
|
|
CHADL
|
chondroadherin-like
|
|
chr9_+_130942029
|
0.898
|
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr17_-_18890963
|
0.890
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr6_-_56824672
|
0.871
|
|
DST
|
dystonin
|
|
chr11_+_63864263
|
0.868
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr6_-_154719560
|
0.850
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr2_-_47986160
|
0.846
|
NM_001190274
|
FBXO11
|
F-box protein 11
|
|
chr5_+_75734759
|
0.810
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr6_-_25044156
|
0.810
|
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr13_-_32678142
|
0.810
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr14_+_99601498
|
0.783
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr1_-_45988901
|
0.778
|
NM_001145349
NM_005897
|
IPP
|
intracisternal A particle-promoted polypeptide
|
|
chrX_-_47374187
|
0.768
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr4_-_25473528
|
0.762
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr19_+_3129735
|
0.762
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr2_-_213723298
|
0.740
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr11_+_59979857
|
0.718
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr15_+_38849450
|
0.705
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr3_+_195889845
|
0.702
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr17_+_64922420
|
0.680
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr17_+_37693090
|
0.677
|
NM_003152
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr11_-_60475558
|
0.670
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr6_-_11490487
|
0.663
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr14_+_99601517
|
0.654
|
|
EVL
|
Enah/Vasp-like
|
|
chr7_+_87401368
|
0.653
|
NM_004194
NM_016351
NM_021721
NM_021722
NM_021723
|
ADAM22
|
ADAM metallopeptidase domain 22
|
|
chr12_+_93066370
|
0.639
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr12_-_8584658
|
0.617
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr11_-_77806500
|
0.609
|
NM_080491
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr11_-_77806413
|
0.605
|
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr7_+_37926943
|
0.603
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr3_-_120761092
|
0.601
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr13_+_107720475
|
0.588
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr10_-_27189898
|
0.587
|
|
ABI1
|
abl-interactor 1
|
|
chr8_+_11388918
|
0.587
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr2_+_149118791
|
0.578
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr6_-_56824371
|
0.578
|
|
DST
|
dystonin
|
|
chr9_+_70509925
|
0.575
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr2_+_65308332
|
0.565
|
NM_001005386
NM_005722
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast)
|
|
chrX_-_100527799
|
0.561
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr5_-_176869463
|
0.560
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr6_-_32268267
|
0.557
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr5_-_176869148
|
0.557
|
|
|
|
|
chr5_+_139485703
|
0.553
|
NM_001007189
|
C5orf53
|
chromosome 5 open reading frame 53
|
|
chr18_+_30875297
|
0.550
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr9_-_115077644
|
0.550
|
NM_139286
|
CDC26
|
cell division cycle 26 homolog (S. cerevisiae)
|
|
chrX_+_51562894
|
0.543
|
NM_001005332
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr2_-_242449662
|
0.539
|
NM_005018
|
PDCD1
|
programmed cell death 1
|
|
chr11_-_58099884
|
0.537
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr10_-_53129315
|
0.534
|
NM_015235
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant
|
|
chr19_-_53558961
|
0.533
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr5_-_176869440
|
0.532
|
|
DOK3
|
docking protein 3
|
|
chr12_+_48765300
|
0.532
|
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr7_-_23476497
|
0.528
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr8_-_120914233
|
0.527
|
NM_003184
|
TAF2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa
|
|
chr16_+_74868676
|
0.525
|
NM_033401
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chrX_+_77267477
|
0.520
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr1_-_156078115
|
0.518
|
NM_005894
|
CD5L
|
CD5 molecule-like
|
|
chr14_+_88130482
|
0.518
|
NM_207662
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr18_-_65766068
|
0.515
|
|
CD226
|
CD226 molecule
|
|
chr12_-_47604930
|
0.514
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr7_+_139175420
|
0.514
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr1_-_181826086
|
0.511
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr17_-_41625921
|
0.509
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr18_-_16945763
|
0.508
|
NM_005406
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr11_+_65026382
|
0.508
|
|
|
|
|
chr11_-_66961709
|
0.506
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr18_-_16945613
|
0.498
|
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr12_-_24628348
|
0.494
|
|
C12orf67
|
chromosome 12 open reading frame 67
|
|
chr11_-_9182356
|
0.489
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr19_-_46550788
|
0.481
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr7_+_23187970
|
0.481
|
NM_007342
|
NUPL2
|
nucleoporin like 2
|
|
chr10_-_103567773
|
0.480
|
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase)
|
|
chr12_-_56422120
|
0.476
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr14_+_61653827
|
0.475
|
|
FLJ43390
|
hypothetical LOC646113
|
|
chr7_-_135312387
|
0.469
|
|
MTPN
|
myotrophin
|
|
chr18_-_51219880
|
0.465
|
|
TCF4
|
transcription factor 4
|
|
chr3_-_122862391
|
0.464
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr2_-_197165495
|
0.464
|
NM_020760
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2
|
|
chr12_+_48443232
|
0.461
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr14_-_21074875
|
0.459
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr12_-_56157898
|
0.456
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr7_+_99793561
|
0.456
|
NM_178238
|
PILRB
|
paired immunoglobin-like type 2 receptor beta
|
|
chrX_-_111809929
|
0.456
|
NM_178175
|
LHFPL1
|
lipoma HMGIC fusion partner-like 1
|
|
chr7_+_149779011
|
0.455
|
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr2_+_233633348
|
0.455
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chrX_-_130792297
|
0.455
|
|
LOC286467
|
hypothetical LOC286467
|
|
chr5_+_176955595
|
0.454
|
|
|
|
|
chr6_-_132875858
|
0.453
|
NM_003569
|
STX7
|
syntaxin 7
|
|
chr6_-_32268615
|
0.451
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr1_-_28287658
|
0.450
|
NM_001990
|
EYA3
|
eyes absent homolog 3 (Drosophila)
|
|
chr7_-_135312574
|
0.449
|
|
MTPN
|
myotrophin
|
|
chr5_-_158567411
|
0.447
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr2_+_233633279
|
0.447
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr15_+_79376295
|
0.446
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr2_-_96174865
|
0.444
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr1_-_27353987
|
0.441
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr17_+_39778191
|
0.441
|
|
GRN
|
granulin
|
|
chr12_+_6930734
|
0.440
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr11_+_86426518
|
0.437
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chrX_-_85189194
|
0.436
|
NM_000390
NM_001145414
|
CHM
|
choroideremia (Rab escort protein 1)
|
|
chr1_+_110031940
|
0.435
|
NM_000561
NM_146421
|
GSTM1
|
glutathione S-transferase mu 1
|
|
chr14_+_49157150
|
0.435
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr3_-_122862447
|
0.434
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr5_-_71838875
|
0.430
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr13_-_32822759
|
0.430
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chrX_-_134257624
|
0.429
|
NM_001185063
NM_007131
|
ZNF75D
|
zinc finger protein 75D
|
|
chr15_+_64466285
|
0.428
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr1_+_12325632
|
0.426
|
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae)
|
|
chr16_-_3567354
|
0.424
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr1_+_151442542
|
0.423
|
NM_001010857
|
LELP1
|
late cornified envelope-like proline-rich 1
|
|
chr12_+_6930778
|
0.420
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr7_+_149778869
|
0.415
|
NM_175571
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr12_+_97433545
|
0.415
|
|
TMPO
|
thymopoietin
|
|
chr7_-_130443379
|
0.415
|
|
FLJ43663
|
hypothetical LOC378805
|
|
chr3_-_15814678
|
0.414
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr7_+_128651976
|
0.413
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr10_+_70553936
|
0.412
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr12_-_104153920
|
0.411
|
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr10_+_70553949
|
0.411
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr15_+_64466161
|
0.410
|
NM_002755
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr3_-_107070400
|
0.408
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr10_+_70553945
|
0.407
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr13_+_107720561
|
0.406
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr4_-_166118267
|
0.406
|
NM_001012414
|
TRIM61
|
tripartite motif containing 61
|
|
chr17_+_55269974
|
0.405
|
|
|
|
|
chr11_-_95715976
|
0.403
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr11_+_298106
|
0.402
|
NM_006435
|
IFITM2
|
interferon induced transmembrane protein 2 (1-8D)
|
|
chr1_-_167969802
|
0.401
|
NM_000450
|
SELE
|
selectin E
|
|
chr12_+_79625538
|
0.401
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr17_-_7061641
|
0.400
|
NM_001128827
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr4_-_101658094
|
0.399
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr12_+_6930694
|
0.398
|
NM_002831
NM_080549
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr4_-_103968104
|
0.398
|
NM_003340
NM_181886
NM_181888
NM_181889
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast)
|
|
chr9_-_115077504
|
0.397
|
|
FKBP15
|
FK506 binding protein 15, 133kDa
|
|
chr17_+_34871775
|
0.396
|
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr10_+_70553862
|
0.396
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chrX_+_70419766
|
0.395
|
NM_001145408
NM_001145409
NM_001145410
NM_007363
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr12_+_45896511
|
0.394
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr5_+_143171918
|
0.393
|
NM_021182
|
HMHB1
|
histocompatibility (minor) HB-1
|
|
chr10_+_70553962
|
0.391
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_+_149638633
|
0.390
|
NM_002796
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4
|
|
chr12_+_6930799
|
0.389
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr8_+_64243741
|
0.389
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr12_+_6930792
|
0.389
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr8_+_64243662
|
0.388
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr17_-_7064092
|
0.388
|
NM_001365
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chrX_-_70248022
|
0.387
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr7_+_149778815
|
0.387
|
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr19_-_7976468
|
0.387
|
NM_001419
|
ELAVL1
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
|
|
chr10_+_91051685
|
0.387
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr5_-_124108672
|
0.387
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr6_+_31647854
|
0.386
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr2_+_154436670
|
0.383
|
NM_052917
|
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13)
|
|
chr10_+_70553993
|
0.383
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr2_+_113648036
|
0.383
|
|
PSD4
|
pleckstrin and Sec7 domain containing 4
|
|
chr12_+_53177758
|
0.383
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr10_+_35524060
|
0.382
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr12_+_6930804
|
0.381
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr11_+_5602782
|
0.381
|
NM_021616
|
TRIM34
|
tripartite motif containing 34
|
|
chr22_-_37425937
|
0.380
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr12_-_131816604
|
0.380
|
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2
|
|
chr10_-_27189995
|
0.379
|
NM_001012750
NM_001012751
NM_001012752
NM_001178116
NM_001178119
NM_001178120
NM_001178121
NM_001178122
NM_001178123
NM_001178124
NM_001178125
NM_005470
|
ABI1
|
abl-interactor 1
|
|
chr14_-_66896167
|
0.379
|
NM_015994
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D
|
|
chr14_+_38720085
|
0.379
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr3_-_130323033
|
0.378
|
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr11_+_4571844
|
0.378
|
NM_001005169
|
OR52I1
|
olfactory receptor, family 52, subfamily I, member 1
|
|
chr10_+_70553983
|
0.376
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_+_202000906
|
0.376
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr12_+_53177789
|
0.376
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr8_-_108579270
|
0.373
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr17_+_18971506
|
0.372
|
NM_001129778
|
GRAPL
|
GRB2-related adaptor protein-like
|
|
chr18_-_59137488
|
0.372
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr12_+_53177806
|
0.371
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr17_-_41018291
|
0.371
|
|
|
|
|
chr1_+_149877955
|
0.371
|
|
SNX27
|
sorting nexin family member 27
|
|
chr11_+_119712113
|
0.370
|
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr12_-_53071285
|
0.370
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr10_+_35524835
|
0.369
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr1_-_200395741
|
0.368
|
NM_080588
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr9_+_126094067
|
0.367
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr15_+_40427592
|
0.366
|
NM_212465
|
CAPN3
|
calpain 3, (p94)
|
|
chr12_+_4253143
|
0.366
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chrX_+_134482387
|
0.366
|
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr17_+_7180571
|
0.365
|
NM_014716
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr3_+_9451506
|
0.363
|
|
|
|
|
chr11_+_125163215
|
0.363
|
NM_001129883
|
PATE3
|
prostate and testis expressed 3
|
|
chr2_+_113648020
|
0.363
|
NM_012455
|
PSD4
|
pleckstrin and Sec7 domain containing 4
|
|
chr3_-_181879955
|
0.362
|
NM_181426
|
CCDC39
|
coiled-coil domain containing 39
|
|
chr16_-_30301357
|
0.362
|
|
SEPT1
|
septin 1
|
|
chr13_+_107719977
|
0.358
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr7_+_23603495
|
0.358
|
NM_138771
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr17_-_38087366
|
0.358
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr3_-_184756101
|
0.355
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|