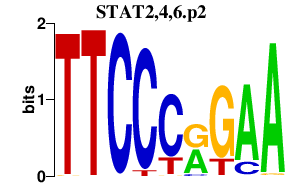
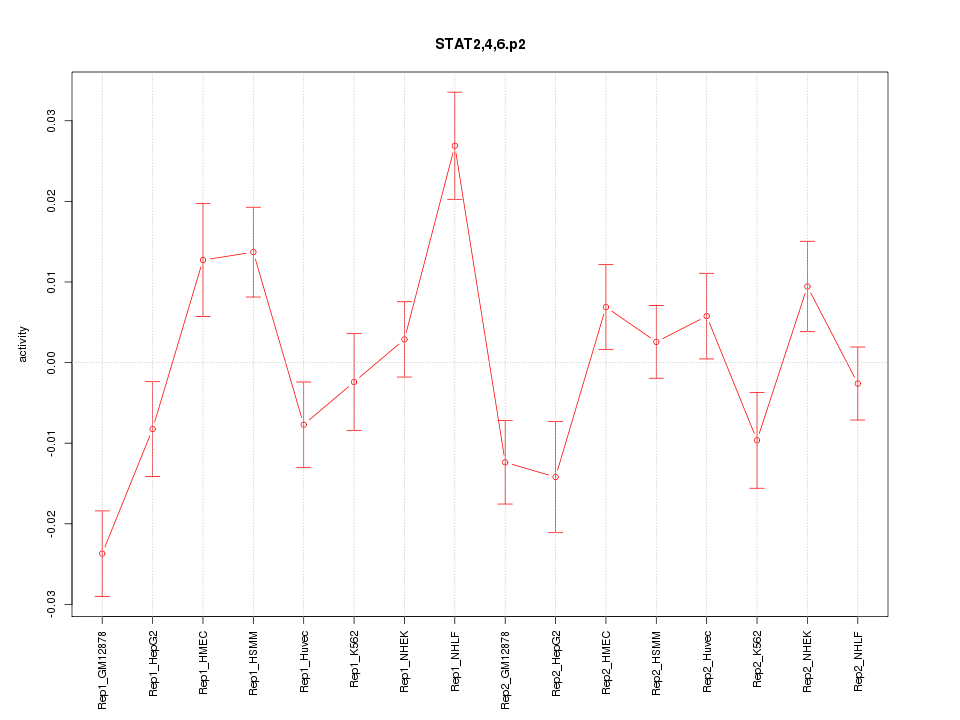
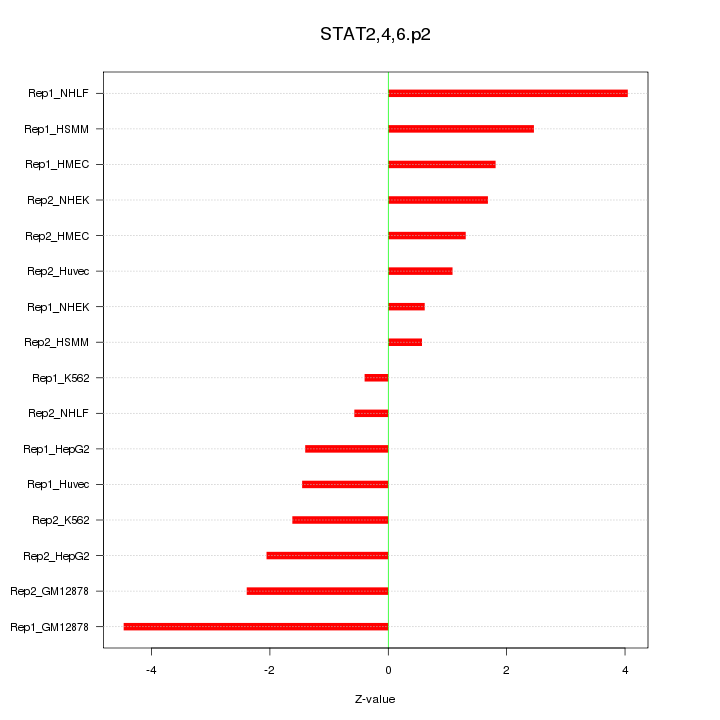
|
chr5_+_140241976
|
3.310
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr1_+_36462583
|
2.517
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr1_+_36462625
|
2.497
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr19_-_49552638
|
2.462
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr12_+_7038240
|
2.422
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr14_+_95792299
|
2.080
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr15_+_68971835
|
1.999
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr12_-_51200347
|
1.981
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr19_+_49248003
|
1.964
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr1_-_171441234
|
1.807
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr16_+_14187965
|
1.743
|
|
MKL2
|
MKL/myocardin-like 2
|
|
chr12_-_55013987
|
1.695
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr10_+_95838801
|
1.643
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr1_+_161305558
|
1.616
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_+_54611212
|
1.566
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr9_+_99109730
|
1.540
|
NM_020893
|
C9orf174
|
chromosome 9 open reading frame 174
|
|
chr3_-_64186151
|
1.461
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr21_-_27139563
|
1.447
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr14_+_23849196
|
1.445
|
NM_001164692
NM_019839
|
LTB4R2
|
leukotriene B4 receptor 2
|
|
chr11_-_82674546
|
1.433
|
|
CCDC90B
|
coiled-coil domain containing 90B
|
|
chr16_-_31390926
|
1.384
|
|
|
|
|
chr10_-_95231933
|
1.375
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr11_+_66546765
|
1.336
|
NM_001177880
|
SYT12
|
synaptotagmin XII
|
|
chr11_-_85115105
|
1.309
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr18_+_59295123
|
1.304
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr6_-_56824672
|
1.289
|
|
DST
|
dystonin
|
|
chr11_+_131286513
|
1.282
|
|
NTM
|
neurotrimin
|
|
chr3_-_42719322
|
1.275
|
NM_020707
|
HHATL
|
hedgehog acyltransferase-like
|
|
chrX_+_105932536
|
1.272
|
NM_017752
NM_198881
|
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain)
|
|
chr5_+_140286485
|
1.242
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr19_+_15079141
|
1.240
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr7_-_5429702
|
1.235
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr2_-_56266408
|
1.220
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr6_+_31190505
|
1.214
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr2_-_38156769
|
1.212
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr19_+_42099073
|
1.205
|
NM_198539
|
ZNF568
|
zinc finger protein 568
|
|
chr19_+_58715988
|
1.198
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr1_+_154178103
|
1.176
|
NM_181885
|
RXFP4
|
relaxin/insulin-like family peptide receptor 4
|
|
chr11_-_82675024
|
1.174
|
NM_021825
|
CCDC90B
|
coiled-coil domain containing 90B
|
|
chr12_-_55013713
|
1.142
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr7_-_83116414
|
1.139
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr2_-_202192141
|
1.136
|
NM_001168216
NM_001168217
NM_001168221
NM_152525
|
ALS2CR11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11
|
|
chr9_+_15412875
|
1.131
|
|
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa
|
|
chr2_+_165858586
|
1.100
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr12_-_68369322
|
1.085
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr20_-_14266200
|
1.075
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr10_-_121345968
|
1.073
|
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr6_-_29081015
|
1.071
|
NM_001010877
|
ZNF311
|
zinc finger protein 311
|
|
chr19_-_42098919
|
1.068
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr6_-_56824371
|
1.053
|
|
DST
|
dystonin
|
|
chr1_-_36589744
|
1.048
|
|
|
|
|
chr12_+_43972732
|
1.023
|
NM_001142678
|
ANO6
|
anoctamin 6
|
|
chr19_+_42400908
|
1.018
|
|
ZNF383
|
zinc finger protein 383
|
|
chr12_+_67366989
|
1.018
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr17_-_8215577
|
1.013
|
NM_213597
|
KRBA2
|
KRAB-A domain containing 2
|
|
chr19_-_42098758
|
1.007
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr6_-_84994032
|
0.995
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr19_+_49361054
|
0.989
|
NM_001146220
NM_001032373
NM_001032374
NM_015919
NM_001032372
|
ZNF226
|
zinc finger protein 226
|
|
chr19_+_54996613
|
0.984
|
|
|
|
|
chr19_-_49309102
|
0.984
|
|
LOC100379224
|
hypothetical LOC100379224
|
|
chr19_+_49023283
|
0.979
|
NM_181845
|
ZNF283
|
zinc finger protein 283
|
|
chr15_-_62435267
|
0.977
|
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr20_+_19903778
|
0.974
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_+_200246312
|
0.966
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr15_-_41185460
|
0.964
|
NM_174916
|
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1
|
|
chr2_-_144994349
|
0.963
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_+_154120170
|
0.954
|
|
SYT11
|
synaptotagmin XI
|
|
chr4_-_122063118
|
0.949
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr1_-_149404954
|
0.943
|
NM_212551
NM_001136543
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1
|
|
chr15_-_68971665
|
0.935
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr3_-_185025989
|
0.934
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr1_-_225572419
|
0.926
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chrX_-_106129854
|
0.923
|
|
MORC4
|
MORC family CW-type zinc finger 4
|
|
chr1_-_151788341
|
0.916
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr9_-_90456824
|
0.915
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr3_+_11153778
|
0.908
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr11_-_113149642
|
0.906
|
|
ZW10
|
ZW10, kinetochore associated, homolog (Drosophila)
|
|
chr8_-_130868315
|
0.904
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr2_-_144994038
|
0.900
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_+_77171938
|
0.899
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr9_-_116920232
|
0.895
|
NM_002160
|
TNC
|
tenascin C
|
|
chr10_+_92621221
|
0.893
|
|
RPP30
|
ribonuclease P/MRP 30kDa subunit
|
|
chr8_-_127639892
|
0.883
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr11_-_113149602
|
0.882
|
NM_004724
|
ZW10
|
ZW10, kinetochore associated, homolog (Drosophila)
|
|
chr21_-_27139143
|
0.862
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr13_-_32010883
|
0.852
|
NM_014887
NM_033111
|
N4BP2L2
|
NEDD4 binding protein 2-like 2
|
|
chr12_+_51730101
|
0.832
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr5_-_102926349
|
0.828
|
NM_031438
|
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12
|
|
chr19_-_1083109
|
0.826
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr8_+_95904762
|
0.825
|
|
INTS8
|
integrator complex subunit 8
|
|
chr3_-_64648370
|
0.820
|
NM_182920
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr9_+_135994734
|
0.817
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr5_-_111120764
|
0.813
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111120817
|
0.812
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_71376919
|
0.806
|
|
FOXP1
|
forkhead box P1
|
|
chr5_+_145807059
|
0.803
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr19_+_60844093
|
0.794
|
NM_207115
|
ZNF580
|
zinc finger protein 580
|
|
chr12_-_83830710
|
0.792
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr6_-_29451042
|
0.791
|
NM_030959
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3
|
|
chr19_-_49500966
|
0.789
|
NM_004234
|
ZNF235
|
zinc finger protein 235
|
|
chr14_+_100365162
|
0.788
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr2_+_33554824
|
0.784
|
NM_001139488
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr8_-_109165012
|
0.778
|
NM_178565
|
RSPO2
|
R-spondin 2 homolog (Xenopus laevis)
|
|
chr1_-_202595666
|
0.774
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr12_-_83830670
|
0.772
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr1_+_68070573
|
0.772
|
|
|
|
|
chr11_-_86061301
|
0.770
|
NM_001161586
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr17_+_7199178
|
0.769
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr12_-_83830700
|
0.768
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr4_-_186934059
|
0.756
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_-_48813256
|
0.753
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr11_-_119513429
|
0.752
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr8_+_67751007
|
0.751
|
NM_019607
|
C8orf44
|
chromosome 8 open reading frame 44
|
|
chr8_+_70567581
|
0.748
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr1_-_68071635
|
0.747
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr3_-_113495763
|
0.744
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr5_+_140733664
|
0.740
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr16_+_31390567
|
0.738
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr9_+_676640
|
0.735
|
|
|
|
|
chr12_-_6356783
|
0.734
|
NM_001159575
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr10_+_74540215
|
0.733
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr11_-_133220578
|
0.732
|
NM_174927
|
SPATA19
|
spermatogenesis associated 19
|
|
chr10_+_92621677
|
0.732
|
NM_001104546
NM_006413
|
RPP30
|
ribonuclease P/MRP 30kDa subunit
|
|
chr2_-_40532651
|
0.731
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr7_-_19123765
|
0.730
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr3_+_116824840
|
0.729
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr8_-_97316933
|
0.727
|
NM_006294
|
UQCRB
|
ubiquinol-cytochrome c reductase binding protein
|
|
chr13_+_26896614
|
0.721
|
NM_002097
|
GTF3A
|
general transcription factor IIIA
|
|
chr20_+_2743613
|
0.715
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr5_-_111121150
|
0.714
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_-_19123787
|
0.712
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr12_+_11693909
|
0.702
|
|
ETV6
|
ets variant 6
|
|
chr4_+_48683021
|
0.701
|
NM_025087
|
CWH43
|
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae)
|
|
chr2_-_38156811
|
0.699
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr3_-_11585264
|
0.692
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr3_-_158700036
|
0.687
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr11_-_118477994
|
0.682
|
NM_001382
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr19_+_43502301
|
0.678
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr2_-_151052391
|
0.675
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr2_-_86186441
|
0.675
|
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr1_+_40769877
|
0.674
|
|
ZNF684
|
zinc finger protein 684
|
|
chr22_-_34349309
|
0.669
|
NM_203377
|
MB
|
myoglobin
|
|
chr17_-_1566223
|
0.667
|
|
C17orf91
|
chromosome 17 open reading frame 91
|
|
chr17_-_7083321
|
0.664
|
NM_024297
|
PHF23
|
PHD finger protein 23
|
|
chrX_+_77041612
|
0.658
|
NM_001866
|
COX7B
|
cytochrome c oxidase subunit VIIb
|
|
chr6_+_54819527
|
0.657
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr1_+_185064654
|
0.657
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr19_+_702108
|
0.654
|
NM_173481
|
C19orf21
|
chromosome 19 open reading frame 21
|
|
chr14_+_52243645
|
0.649
|
NM_002806
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6
|
|
chr17_+_36515166
|
0.648
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr19_+_49309350
|
0.648
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr6_+_129245978
|
0.647
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr2_-_31215074
|
0.646
|
NM_024572
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr5_-_138238877
|
0.641
|
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr12_-_119172339
|
0.639
|
NM_025157
|
PXN
|
paxillin
|
|
chr2_-_61551403
|
0.639
|
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr2_-_175255717
|
0.639
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr8_-_127639647
|
0.637
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr15_+_72397934
|
0.636
|
NM_182791
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr4_+_77575276
|
0.636
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr17_+_1566566
|
0.635
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr19_-_39859997
|
0.633
|
|
|
|
|
chr3_+_98966253
|
0.633
|
NM_032146
NM_177976
|
ARL6
|
ADP-ribosylation factor-like 6
|
|
chr11_-_27637771
|
0.633
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr8_+_106400322
|
0.630
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr10_-_121346464
|
0.628
|
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr1_+_154520693
|
0.627
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr7_+_110518297
|
0.626
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr2_+_159533329
|
0.624
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr14_+_64523700
|
0.624
|
|
|
|
|
chr19_+_43502380
|
0.624
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr1_+_200698493
|
0.623
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr1_+_207823667
|
0.622
|
NM_020439
|
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG
|
|
chr5_+_38882017
|
0.620
|
|
OSMR
|
oncostatin M receptor
|
|
chr16_+_4485859
|
0.619
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr4_-_155731288
|
0.618
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr10_+_88506320
|
0.616
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr1_+_40769819
|
0.616
|
NM_152373
|
ZNF684
|
zinc finger protein 684
|
|
chr6_+_108593907
|
0.616
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr17_-_44047299
|
0.615
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr20_+_5679032
|
0.610
|
NM_152504
|
C20orf196
|
chromosome 20 open reading frame 196
|
|
chr17_+_29606408
|
0.607
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr1_+_200246370
|
0.607
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr4_-_89963177
|
0.599
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr10_-_121346517
|
0.599
|
NM_001033925
NM_003252
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr8_-_144313100
|
0.599
|
NM_001135655
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr12_-_87498225
|
0.599
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr15_-_53440069
|
0.597
|
|
CCPG1
|
cell cycle progression 1
|
|
chr2_-_218516950
|
0.596
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr9_-_133944877
|
0.596
|
|
MED27
|
mediator complex subunit 27
|
|
chr12_+_54904417
|
0.595
|
|
OBFC2B
|
oligonucleotide/oligosaccharide-binding fold containing 2B
|
|
chr3_+_173043832
|
0.595
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr5_-_92982927
|
0.583
|
|
|
|
|
chr17_+_16283025
|
0.583
|
|
NCRNA00188
|
non-protein coding RNA 188
|
|
chr15_-_62435412
|
0.581
|
NM_022048
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr20_+_11846536
|
0.581
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr3_+_50204026
|
0.579
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr14_+_74815233
|
0.575
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr18_-_63333487
|
0.569
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr17_-_4841628
|
0.566
|
NM_001167985
NM_001167986
NM_001167987
NM_213726
|
INCA1
|
inhibitor of CDK, cyclin A1 interacting protein 1
|