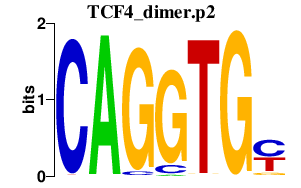
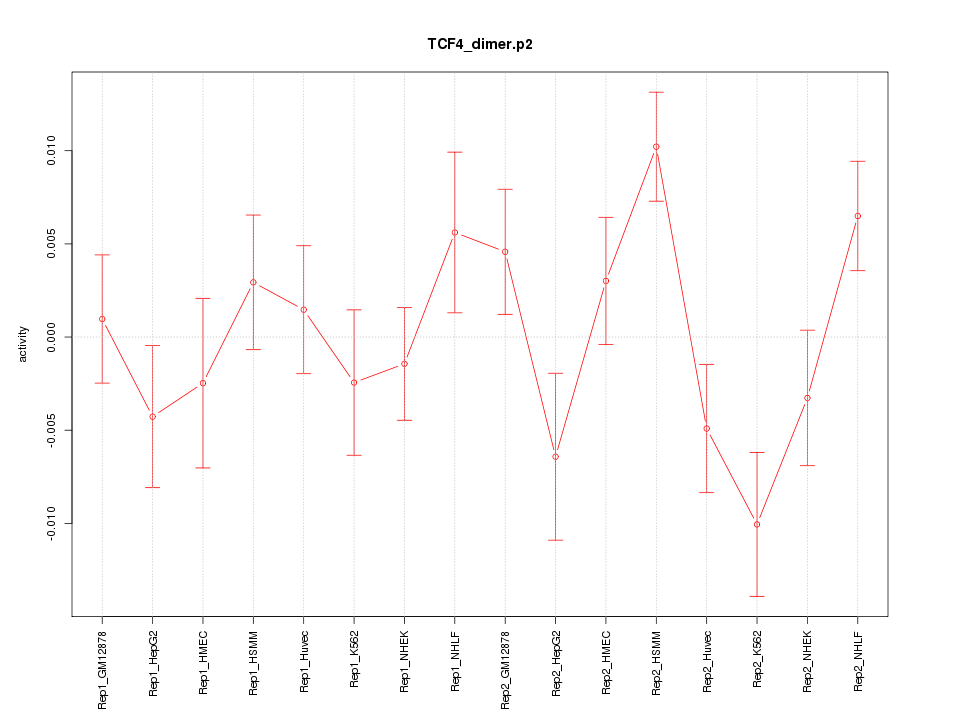
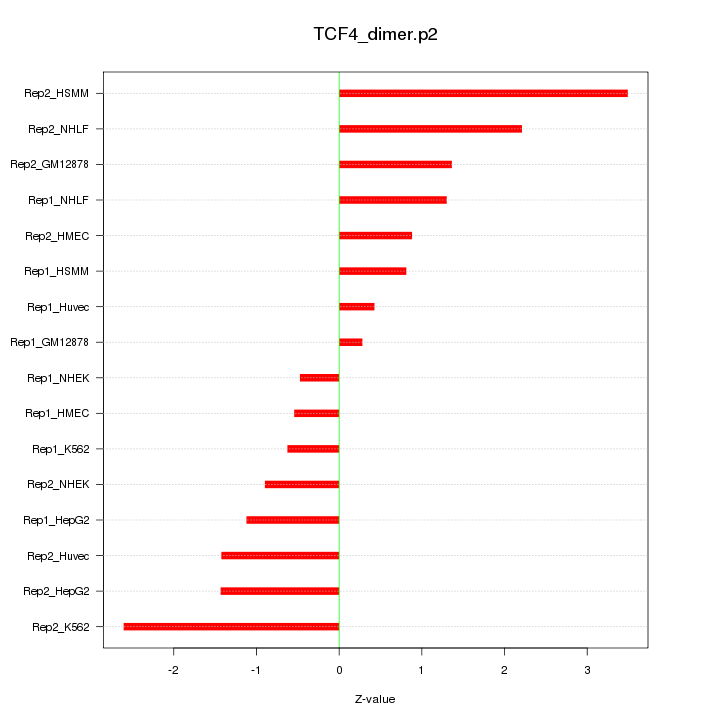
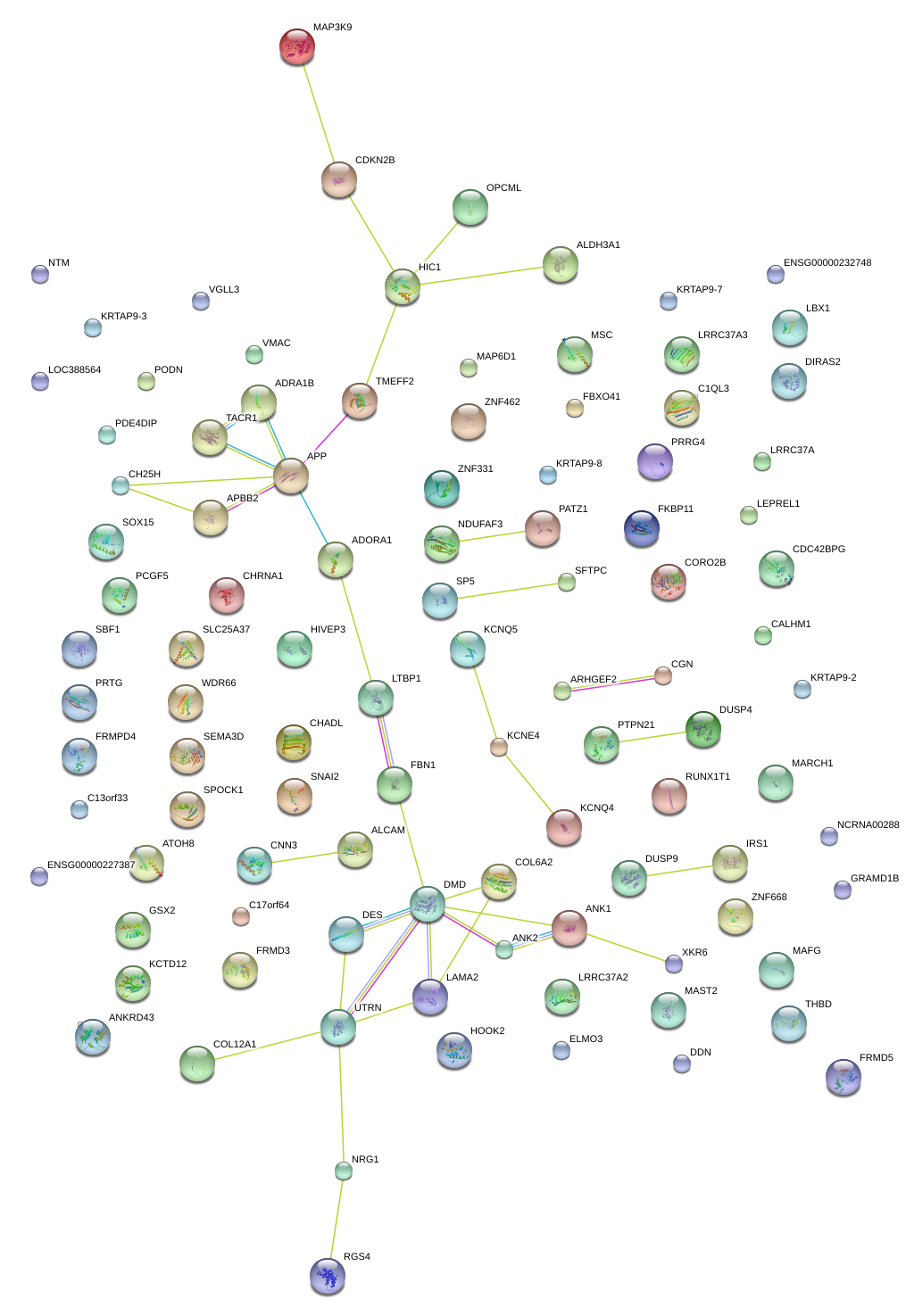
|
chr15_-_42274760
|
2.202
|
|
FRMD5
|
FERM domain containing 5
|
|
chr15_-_42274529
|
2.151
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr19_-_12747263
|
1.636
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr1_-_143643064
|
1.545
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_+_201364050
|
1.542
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr2_+_219991342
|
1.540
|
NM_001927
|
DES
|
desmin
|
|
chr3_+_49034050
|
1.489
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chrX_+_12066505
|
1.473
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr3_+_49034494
|
1.455
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr11_+_131286513
|
1.420
|
|
NTM
|
neurotrimin
|
|
chr3_-_87122936
|
1.403
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr10_-_16604009
|
1.381
|
NM_001010908
|
C1QL3
|
complement component 1, q subcomponent-like 3
|
|
chr4_+_114433551
|
1.362
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr5_+_159276317
|
1.342
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr17_-_7434113
|
1.331
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr9_-_21999270
|
1.287
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr5_-_136862130
|
1.263
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr19_-_12747421
|
1.256
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr8_-_93099040
|
1.233
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_+_54660954
|
1.227
|
NM_133267
|
GSX2
|
GS homeobox 2
|
|
chr8_-_72918697
|
1.223
|
|
MSC
|
musculin
|
|
chr10_-_102978678
|
1.205
|
NM_006562
|
LBX1
|
ladybird homeobox 1
|
|
chr16_+_65790514
|
1.190
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr1_+_46151845
|
1.189
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr1_-_32479785
|
1.167
|
|
MTMR9LP
|
myotubularin related protein 9-like, pseudogene
|
|
chr8_-_72918938
|
1.155
|
|
MSC
|
musculin
|
|
chr8_+_72918772
|
1.150
|
|
LOC100132891
|
hypothetical LOC100132891
|
|
chr2_+_33213111
|
1.131
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr17_+_36636404
|
1.130
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr3_+_106568334
|
1.129
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr2_+_85834111
|
1.126
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr8_+_72918889
|
1.120
|
|
|
|
|
chr17_+_1905104
|
1.105
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr5_-_136862317
|
1.100
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr17_-_19589363
|
1.098
|
NM_001135168
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr1_+_161305019
|
1.098
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_-_47604930
|
1.092
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr9_-_92444886
|
1.078
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr3_+_106568243
|
1.077
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr13_-_76358370
|
1.060
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr2_-_75280121
|
1.050
|
NM_001058
NM_015727
|
TACR1
|
tachykinin receptor 1
|
|
chr15_-_46725087
|
1.048
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr16_-_30983909
|
1.038
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr3_-_87122958
|
1.030
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr17_-_60345364
|
1.020
|
NM_199340
|
LRRC37A3
|
leucine rich repeat containing 37, member A3
|
|
chr3_+_106568430
|
0.991
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr11_+_126378507
|
0.990
|
|
NCRNA00288
|
non-protein coding RNA 288
|
|
chr17_-_19589516
|
0.987
|
NM_001135167
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr21_+_46355797
|
0.982
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr13_+_30378311
|
0.978
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr2_-_73350103
|
0.961
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr7_-_84654106
|
0.960
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr8_-_49996353
|
0.956
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr11_+_122936145
|
0.955
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr11_+_32808045
|
0.948
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr17_+_55854636
|
0.935
|
NM_181707
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr8_-_11096257
|
0.922
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr19_+_58750305
|
0.912
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr2_+_171280944
|
0.903
|
|
SP5
|
Sp5 transcription factor
|
|
chr12_-_47679176
|
0.892
|
NM_015086
|
DDN
|
dendrin
|
|
chr1_+_161305188
|
0.888
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr22_-_39964001
|
0.876
|
|
CHADL
|
chondroadherin-like
|
|
chr15_-_53822468
|
0.869
|
NM_173814
|
PRTG
|
protogenin
|
|
chr1_-_153107084
|
0.866
|
|
|
|
|
chr1_+_149750485
|
0.852
|
NM_020770
|
CGN
|
cingulin
|
|
chr6_-_75972473
|
0.852
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chrX_-_33267646
|
0.851
|
NM_000109
|
DMD
|
dystrophin
|
|
chr12_+_120840829
|
0.848
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr2_-_192767494
|
0.803
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr4_-_165523856
|
0.802
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr15_+_66658626
|
0.801
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr22_-_30071565
|
0.794
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr19_-_60587414
|
0.793
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr6_+_129245978
|
0.786
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr3_-_185025989
|
0.785
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr9_-_85342868
|
0.785
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr4_-_40911212
|
0.782
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr1_-_143643324
|
0.774
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr8_-_29262240
|
0.767
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr2_+_223625105
|
0.765
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr6_-_75972286
|
0.758
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr2_-_175337386
|
0.758
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr17_+_36647795
|
0.758
|
NM_031963
|
KRTAP9-8
|
keratin associated protein 9-8
|
|
chr2_-_227371698
|
0.755
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr14_-_70345485
|
0.751
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr19_+_5855842
|
0.749
|
NM_001017921
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein
|
|
chr9_+_108665168
|
0.746
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr8_+_32525269
|
0.743
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr14_-_88087575
|
0.736
|
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chrX_+_152562013
|
0.724
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr22_-_30071761
|
0.723
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr1_+_53300410
|
0.720
|
NM_153703
|
PODN
|
podocan
|
|
chr20_-_22978141
|
0.717
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr21_-_26464790
|
0.714
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr17_-_77474597
|
0.714
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr6_+_73388529
|
0.711
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr22_-_49240594
|
0.706
|
|
SBF1
|
SET binding factor 1
|
|
chr3_+_50272648
|
0.705
|
|
|
|
|
chr1_-_95165089
|
0.698
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr4_+_114190233
|
0.692
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_-_191321601
|
0.692
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr10_-_90957028
|
0.690
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr10_+_92970302
|
0.690
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr1_-_42157077
|
0.679
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr8_-_41641881
|
0.678
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr10_-_105208634
|
0.677
|
NM_001001412
|
CALHM1
|
calcium homeostasis modulator 1
|
|
chr3_-_191321367
|
0.676
|
|
LEPREL1
|
leprecan-like 1
|
|
chr1_-_154205675
|
0.674
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr5_+_132176921
|
0.671
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr11_-_64368616
|
0.671
|
NM_017525
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like)
|
|
chr17_+_36515166
|
0.666
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr19_+_18608827
|
0.664
|
NM_018316
|
KLHL26
|
kelch-like 26 (Drosophila)
|
|
chr12_-_46585032
|
0.663
|
NM_000376
NM_001017535
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr1_-_42156957
|
0.662
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr5_+_110587546
|
0.660
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr6_-_46091238
|
0.656
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr11_+_46325457
|
0.653
|
NM_003646
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chrX_-_135941498
|
0.644
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr1_-_84928473
|
0.642
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chrX_-_154341451
|
0.641
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr3_-_64186151
|
0.638
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr12_-_107745455
|
0.637
|
NM_001161331
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr3_-_11598829
|
0.631
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr6_+_46205659
|
0.631
|
NM_014936
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative)
|
|
chr1_+_238321794
|
0.631
|
NM_020066
|
FMN2
|
formin 2
|
|
chr15_-_49184764
|
0.630
|
NM_207381
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr4_-_78038025
|
0.629
|
NM_001029870
|
ANKRD56
|
ankyrin repeat domain 56
|
|
chr4_+_129951877
|
0.627
|
|
PHF17
|
PHD finger protein 17
|
|
chr6_+_137285071
|
0.625
|
NM_001008783
|
SLC35D3
|
solute carrier family 35, member D3
|
|
chr15_-_72981595
|
0.624
|
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chr7_-_124192265
|
0.623
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr14_+_61653827
|
0.621
|
|
FLJ43390
|
hypothetical LOC646113
|
|
chr5_+_167651063
|
0.617
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr9_+_99785465
|
0.616
|
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B
|
|
chr9_+_99785405
|
0.614
|
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B
|
|
chr22_-_30072217
|
0.614
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr1_-_203320153
|
0.610
|
NM_203376
|
TMEM81
|
transmembrane protein 81
|
|
chr1_-_42156913
|
0.608
|
|
|
|
|
chr12_-_50871953
|
0.606
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr19_-_55008172
|
0.604
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr8_+_121206478
|
0.603
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr10_+_92970487
|
0.603
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr8_-_17315171
|
0.602
|
|
MTMR7
|
myotubularin related protein 7
|
|
chr9_+_99785277
|
0.596
|
NM_006401
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B
|
|
chr17_+_65677255
|
0.596
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chrX_+_48221323
|
0.595
|
NM_177434
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr8_+_143805595
|
0.594
|
NM_016647
|
C8orf55
|
chromosome 8 open reading frame 55
|
|
chr11_-_17366781
|
0.590
|
NM_000525
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr3_-_73756668
|
0.590
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr11_-_129803515
|
0.583
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr18_+_17076166
|
0.583
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr3_-_198767202
|
0.582
|
NM_004051
NM_203314
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr6_+_73388713
|
0.580
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr6_+_46205777
|
0.579
|
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative)
|
|
chr19_+_1022208
|
0.575
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr12_-_21985433
|
0.573
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr4_+_37569093
|
0.570
|
NM_015173
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr4_+_151218862
|
0.568
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr7_-_47588652
|
0.568
|
|
TNS3
|
tensin 3
|
|
chr1_+_95058687
|
0.565
|
NM_152369
|
SLC44A3
|
solute carrier family 44, member 3
|
|
chr3_-_69254226
|
0.563
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr15_+_90198567
|
0.563
|
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1
|
|
chr6_-_132764257
|
0.559
|
NM_015529
|
MOXD1
|
monooxygenase, DBH-like 1
|
|
chr3_+_120904555
|
0.556
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr22_-_22511173
|
0.556
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr1_+_144273662
|
0.555
|
|
ANKRD35
|
ankyrin repeat domain 35
|
|
chr22_+_29122932
|
0.553
|
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chrX_-_137621492
|
0.544
|
NM_004114
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_-_152185764
|
0.543
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr11_+_113815317
|
0.541
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr15_+_72397934
|
0.541
|
NM_182791
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr9_+_99785493
|
0.540
|
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B
|
|
chr22_+_36401558
|
0.536
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr10_-_99248355
|
0.530
|
NM_022362
|
MMS19
|
MMS19 nucleotide excision repair homolog (S. cerevisiae)
|
|
chr1_-_92721633
|
0.530
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr19_-_11352850
|
0.526
|
|
EPOR
|
erythropoietin receptor
|
|
chr16_-_63713485
|
0.523
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr10_+_31648103
|
0.522
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr7_+_6596183
|
0.522
|
NM_024067
|
C7orf26
|
chromosome 7 open reading frame 26
|
|
chr19_-_2378581
|
0.521
|
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr1_+_95058781
|
0.519
|
|
SLC44A3
|
solute carrier family 44, member 3
|
|
chr7_+_115953582
|
0.517
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_-_107089387
|
0.517
|
|
LOC286002
|
hypothetical LOC286002
|
|
chr10_+_116843108
|
0.516
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr17_+_4922445
|
0.511
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr11_-_70641267
|
0.507
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr12_-_78608154
|
0.505
|
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr8_+_22492586
|
0.503
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr1_-_160260210
|
0.502
|
NM_015441
|
OLFML2B
|
olfactomedin-like 2B
|
|
chr15_-_23659329
|
0.498
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr11_+_65536037
|
0.497
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr10_-_124595680
|
0.497
|
NM_022034
|
CUZD1
|
CUB and zona pellucida-like domains 1
|
|
chr16_-_63713466
|
0.495
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chrX_-_74062005
|
0.495
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr10_+_99463446
|
0.495
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr2_-_208342296
|
0.494
|
NM_003468
|
FZD5
|
frizzled homolog 5 (Drosophila)
|
|
chr10_-_31647925
|
0.494
|
|
LOC220930
|
hypothetical LOC220930
|
|
chr2_+_209996913
|
0.494
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr9_-_74757735
|
0.493
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr1_-_3702908
|
0.493
|
NM_020710
|
LRRC47
|
leucine rich repeat containing 47
|