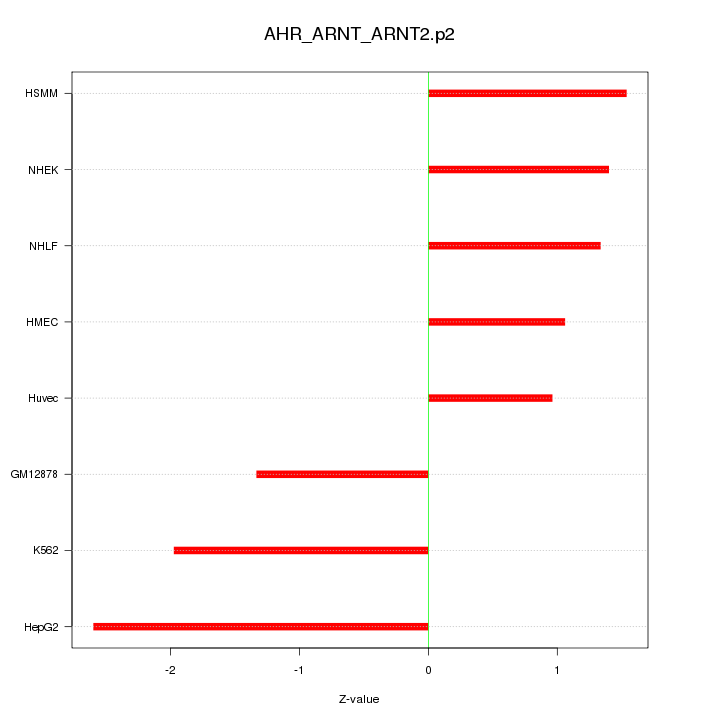
|
chr21_-_38954433
|
4.075
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr11_-_6298284
|
3.505
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr3_-_162305319
|
3.265
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr1_+_110494654
|
2.769
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr13_-_77391525
|
2.650
|
|
|
|
|
chr9_+_4480193
|
2.545
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr18_+_19523526
|
2.334
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr9_-_25668230
|
2.290
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr4_+_81337663
|
2.186
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr3_+_135996990
|
2.167
|
|
EPHB1
|
EPH receptor B1
|
|
chr2_+_29191711
|
2.090
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr19_-_61680510
|
2.068
|
NM_022103
|
ZNF667
|
zinc finger protein 667
|
|
chr11_-_78829342
|
2.055
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr22_-_17891777
|
2.036
|
|
CLDN5
|
claudin 5
|
|
chr6_-_40663089
|
2.021
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr3_-_129024665
|
1.975
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr13_-_37341859
|
1.965
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr13_-_32822759
|
1.957
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr9_-_112381593
|
1.911
|
NM_153366
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr4_+_94969064
|
1.894
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chr1_+_199519202
|
1.881
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr16_+_81218142
|
1.848
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr2_-_45090025
|
1.823
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr3_+_135996734
|
1.779
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr9_-_34579679
|
1.740
|
NM_001842
NM_147164
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr15_+_81906983
|
1.682
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr8_+_31616809
|
1.646
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr10_+_105334704
|
1.630
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr16_+_81218074
|
1.629
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr8_-_6680429
|
1.612
|
NM_207411
|
XKR5
|
XK, Kell blood group complex subunit-related family, member 5
|
|
chr15_+_81907176
|
1.603
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr12_+_58276114
|
1.600
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr15_-_73719546
|
1.598
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr8_+_120497732
|
1.588
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr5_-_159672060
|
1.585
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr5_+_42459568
|
1.571
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chrX_-_83329562
|
1.542
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr7_-_131911768
|
1.533
|
|
PLXNA4
|
plexin A4
|
|
chr14_-_53491019
|
1.523
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr1_+_77520874
|
1.514
|
NM_012093
|
AK5
|
adenylate kinase 5
|
|
chr4_-_57671112
|
1.496
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr2_-_40532651
|
1.478
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr4_-_143987053
|
1.478
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chrX_-_54401161
|
1.478
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chrX_-_107866262
|
1.460
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr3_-_62334229
|
1.437
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chr1_+_32703225
|
1.428
|
NM_001145720
|
ZBTB8B
|
zinc finger and BTB domain containing 8B
|
|
chr5_-_168660293
|
1.424
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr18_-_33399997
|
1.411
|
NM_001025087
NM_001025088
NM_001025089
NM_020180
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr20_+_13924014
|
1.398
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr7_-_27136876
|
1.393
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr7_+_113513828
|
1.383
|
|
FOXP2
|
forkhead box P2
|
|
chr16_+_22733307
|
1.369
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr20_+_13924263
|
1.340
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr12_-_70953461
|
1.330
|
|
LOC283392
|
hypothetical LOC283392
|
|
chr9_+_86473273
|
1.320
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr9_-_23816062
|
1.306
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr6_-_48144344
|
1.285
|
NM_001013732
NM_207499
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr6_+_84619703
|
1.273
|
NM_001009994
|
RIPPLY2
|
ripply2 homolog (zebrafish)
|
|
chr4_-_147079410
|
1.271
|
|
|
|
|
chr11_+_128266460
|
1.255
|
NM_000890
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5
|
|
chr16_-_85099941
|
1.249
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr4_-_122063118
|
1.226
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr4_-_57671273
|
1.221
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr20_+_41978195
|
1.220
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr19_+_38804694
|
1.215
|
NM_001127895
NM_001127896
|
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8
|
|
chr7_-_27149750
|
1.205
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr16_-_86082818
|
1.199
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr10_-_128884411
|
1.190
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chr1_+_77520315
|
1.188
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr4_-_57671307
|
1.187
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr4_-_108176901
|
1.183
|
NM_014421
|
DKK2
|
dickkopf homolog 2 (Xenopus laevis)
|
|
chr5_-_134942346
|
1.161
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr2_+_219141546
|
1.159
|
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr7_-_131911808
|
1.157
|
NM_001105543
NM_020911
|
PLXNA4
|
plexin A4
|
|
chr4_+_156808261
|
1.155
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr9_-_85342868
|
1.146
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr5_-_107034484
|
1.140
|
NM_001962
|
EFNA5
|
ephrin-A5
|
|
chr1_+_77520190
|
1.137
|
|
AK5
|
adenylate kinase 5
|
|
chr5_-_134942867
|
1.134
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chrX_+_21302317
|
1.117
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr8_-_26427304
|
1.089
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr6_+_137285071
|
1.084
|
NM_001008783
|
SLC35D3
|
solute carrier family 35, member D3
|
|
chr6_+_1336077
|
1.082
|
|
|
|
|
chr18_-_24011349
|
1.082
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr10_+_105243724
|
1.079
|
NM_004210
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr22_-_17892001
|
1.079
|
|
CLDN5
|
claudin 5
|
|
chr14_+_60045621
|
1.075
|
NM_007374
|
SIX6
|
SIX homeobox 6
|
|
chr8_-_24869945
|
1.072
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr3_-_135231417
|
1.061
|
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr14_-_73106243
|
1.060
|
|
|
|
|
chr3_+_64645819
|
1.056
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr7_+_28415495
|
1.049
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr5_+_32747194
|
1.049
|
NM_000908
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr9_-_83494198
|
1.037
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr7_-_27120015
|
1.035
|
|
HOXA3
|
homeobox A3
|
|
chr7_+_113513726
|
1.035
|
|
FOXP2
|
forkhead box P2
|
|
chr2_+_219141761
|
1.023
|
NM_005444
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr10_-_49483002
|
1.009
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr17_-_16413188
|
1.005
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr4_-_115120251
|
0.998
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr18_-_33399601
|
0.997
|
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr16_+_22732982
|
0.991
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr15_-_61461127
|
0.985
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr7_+_113513600
|
0.985
|
|
FOXP2
|
forkhead box P2
|
|
chr12_-_22378833
|
0.983
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr4_-_55686503
|
0.977
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr12_-_87498225
|
0.976
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr2_-_213111525
|
0.963
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr10_+_102782151
|
0.962
|
|
SFXN3
|
sideroflexin 3
|
|
chr13_-_77390907
|
0.962
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr2_+_206255376
|
0.961
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr6_+_29799095
|
0.960
|
NM_001098478
NM_001098479
NM_018950
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr22_+_40702709
|
0.958
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr6_+_108593907
|
0.955
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr9_+_86474553
|
0.944
|
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr8_+_59069522
|
0.941
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr3_-_65999548
|
0.936
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr10_+_101079099
|
0.936
|
|
CNNM1
|
cyclin M1
|
|
chr16_+_65790514
|
0.932
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr10_+_80672842
|
0.932
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr10_+_124210980
|
0.932
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr1_+_182040837
|
0.931
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr1_-_58815415
|
0.922
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr15_-_61460967
|
0.921
|
|
CA12
|
carbonic anhydrase XII
|
|
chr17_-_72045218
|
0.921
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr4_-_147079018
|
0.914
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chrX_+_37315934
|
0.914
|
|
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial)
|
|
chr5_-_146238336
|
0.904
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr18_+_40514772
|
0.893
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr14_+_73105618
|
0.892
|
|
ACOT2
|
acyl-CoA thioesterase 2
|
|
chr9_-_78710698
|
0.888
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr14_-_23850481
|
0.880
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr5_+_80292313
|
0.874
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr15_-_77169894
|
0.871
|
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr14_+_23850494
|
0.868
|
NM_001143919
|
LTB4R
|
leukotriene B4 receptor
|
|
chr14_-_23850410
|
0.867
|
NM_014430
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr15_-_43602293
|
0.867
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr17_-_60208083
|
0.865
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr22_-_36153449
|
0.859
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr18_+_48121067
|
0.858
|
|
DCC
|
deleted in colorectal carcinoma
|
|
chr20_+_51023159
|
0.852
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr5_-_83715947
|
0.839
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr6_+_35290160
|
0.828
|
NM_152753
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr14_-_23850466
|
0.828
|
|
|
|
|
chr6_+_133604203
|
0.827
|
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr11_+_66547391
|
0.823
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chr5_-_83716346
|
0.820
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr4_-_17392232
|
0.815
|
NM_015688
|
FAM184B
|
family with sequence similarity 184, member B
|
|
chr4_-_168392151
|
0.814
|
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr15_-_80123935
|
0.813
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr1_+_208472817
|
0.812
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr10_+_102782156
|
0.810
|
|
SFXN3
|
sideroflexin 3
|
|
chr15_-_61680003
|
0.805
|
|
LOC100130855
|
hypothetical LOC100130855
|
|
chr4_-_168392307
|
0.804
|
NM_001040159
NM_016950
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr6_+_133604181
|
0.801
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr7_-_45927263
|
0.796
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr4_+_158361185
|
0.788
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr10_+_12431734
|
0.783
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr15_+_67009892
|
0.778
|
NM_145658
NM_001184780
|
SPESP1
NOX5
|
sperm equatorial segment protein 1
NADPH oxidase, EF-hand calcium binding domain 5
|
|
chr15_+_72006015
|
0.776
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr11_+_66827563
|
0.775
|
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr20_+_51022352
|
0.773
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr9_+_86474445
|
0.773
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr7_+_98084531
|
0.771
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr18_-_5533967
|
0.768
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr12_-_80676562
|
0.768
|
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr4_-_113656762
|
0.766
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr8_-_125809831
|
0.765
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr18_+_19523402
|
0.762
|
|
LAMA3
|
laminin, alpha 3
|
|
chr8_-_38245694
|
0.761
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr5_-_172688024
|
0.755
|
|
STC2
|
stanniocalcin 2
|
|
chr7_-_15692551
|
0.748
|
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr7_-_84654106
|
0.746
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr8_+_1759502
|
0.744
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr3_-_129023850
|
0.744
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chrX_+_129133234
|
0.743
|
NM_004794
|
RAB33A
|
RAB33A, member RAS oncogene family
|
|
chr19_-_2378581
|
0.743
|
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr17_-_8000362
|
0.742
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr8_-_97242119
|
0.741
|
NM_001001557
|
GDF6
|
growth differentiation factor 6
|
|
chr13_-_35603513
|
0.740
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr19_+_52215982
|
0.740
|
NM_002517
|
NPAS1
|
neuronal PAS domain protein 1
|
|
chr6_+_73388291
|
0.740
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr17_-_959007
|
0.738
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr20_-_56523251
|
0.738
|
NM_153360
|
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like
|
|
chr1_+_32992131
|
0.735
|
|
|
|
|
chr13_-_43259032
|
0.729
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr14_+_73073570
|
0.727
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chrX_-_135941498
|
0.726
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr3_-_191321367
|
0.725
|
|
LEPREL1
|
leprecan-like 1
|
|
chr5_+_63497408
|
0.721
|
NM_001113561
NM_178532
|
RNF180
|
ring finger protein 180
|
|
chr22_-_44828636
|
0.720
|
NM_018280
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr13_-_35603466
|
0.720
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr13_-_35603432
|
0.720
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr11_+_66827494
|
0.718
|
NM_017857
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr10_+_59942874
|
0.717
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr19_-_2378530
|
0.716
|
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr8_+_69405510
|
0.712
|
NM_001195639
NM_052958
|
C8orf34
|
chromosome 8 open reading frame 34
|
|
chr12_+_70952729
|
0.711
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr2_-_39040981
|
0.711
|
|
LOC375196
|
hypothetical LOC375196
|