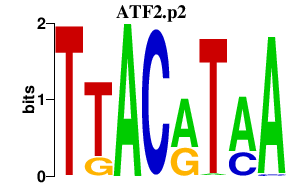
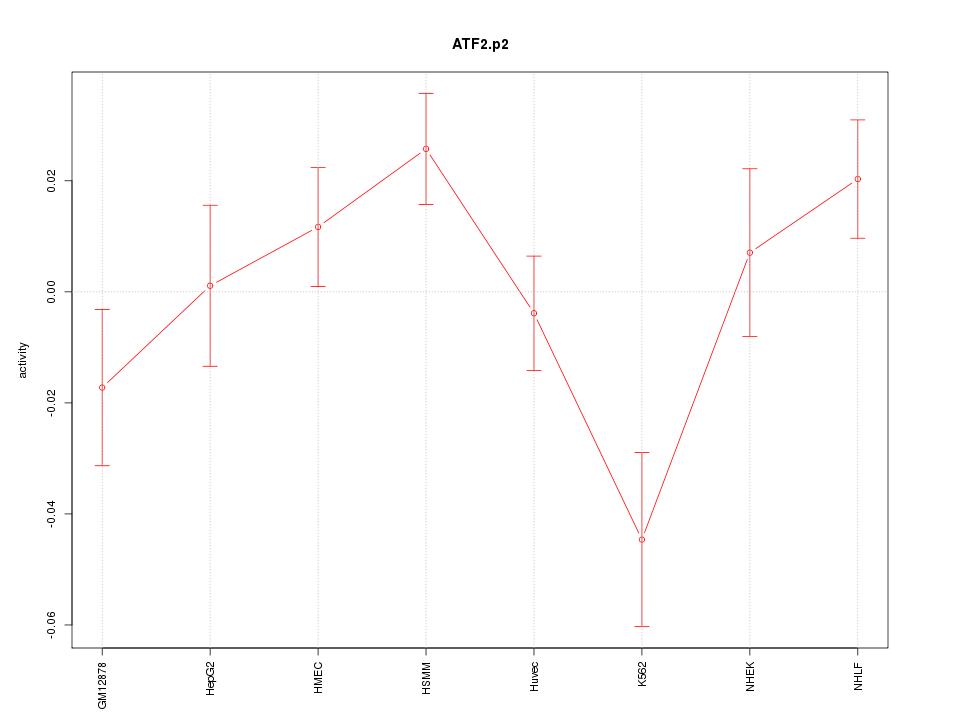
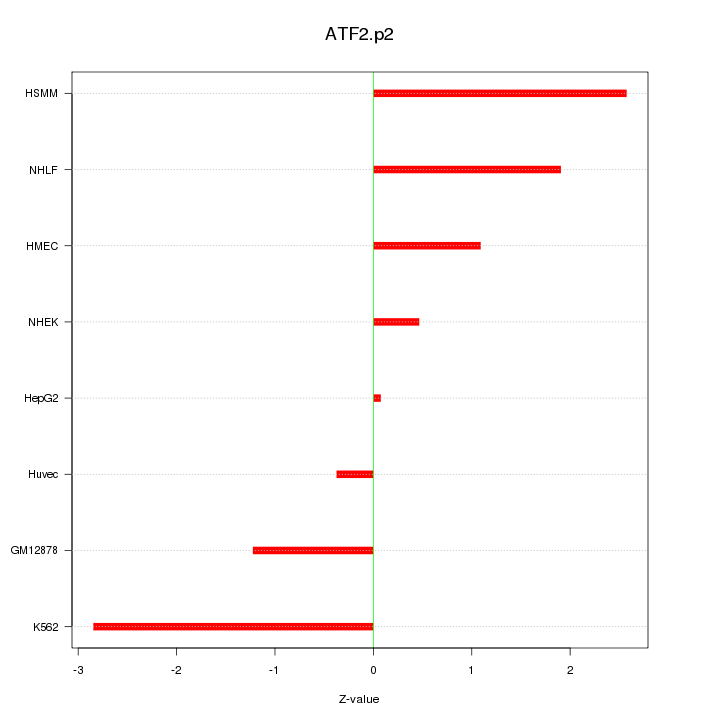
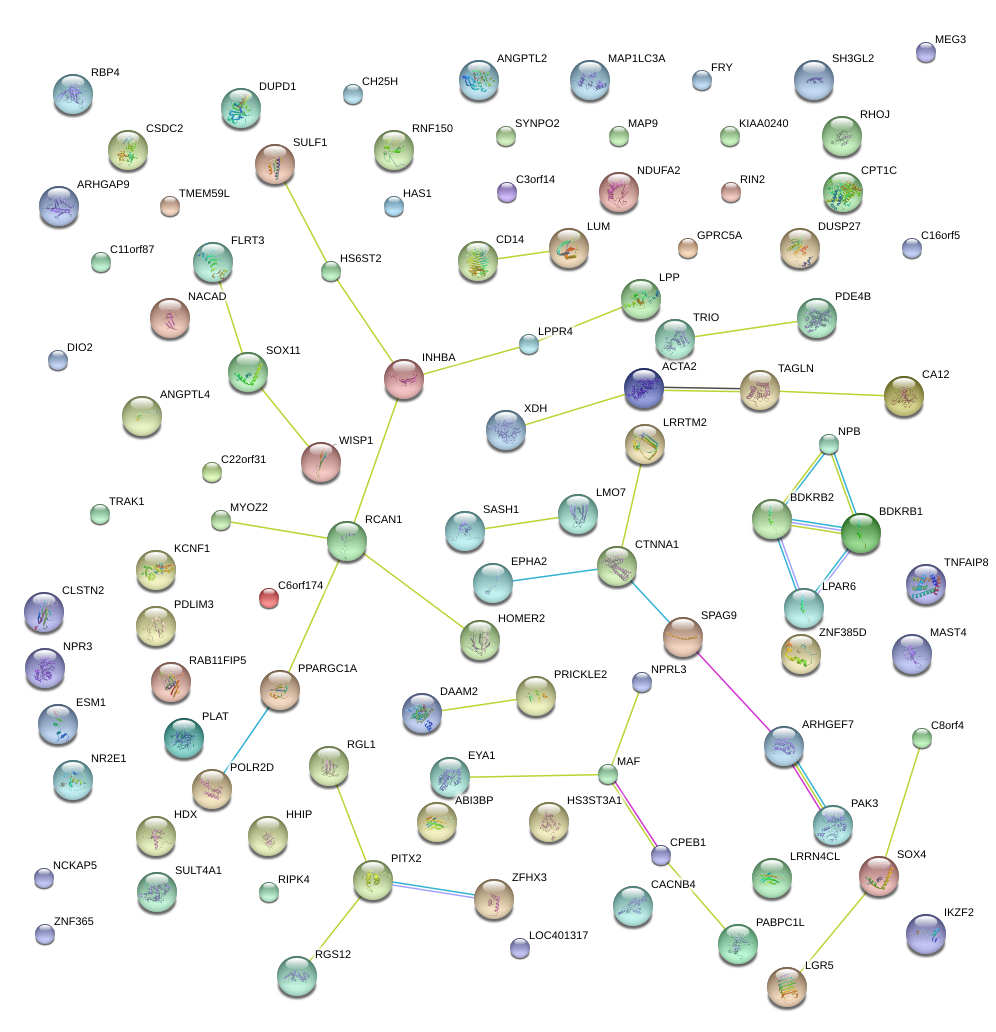
|
chr4_-_23500706
|
2.900
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr8_+_70567634
|
2.352
|
|
SULF1
|
sulfatase 1
|
|
chr3_-_21767819
|
2.292
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr11_+_116575249
|
2.270
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr12_-_123568781
|
2.170
|
|
|
|
|
chr3_+_62279707
|
2.111
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr8_+_70567581
|
2.058
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr12_+_70119785
|
2.038
|
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr16_-_78192111
|
1.984
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr6_-_127822227
|
1.973
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr6_+_39868116
|
1.947
|
|
|
|
|
chr14_+_95740916
|
1.899
|
|
BDKRB2
|
bradykinin receptor B2
|
|
chr7_-_41709191
|
1.896
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr2_+_10969513
|
1.865
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr3_-_64186151
|
1.865
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr3_+_189354167
|
1.850
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr6_+_39868126
|
1.820
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr4_-_186693574
|
1.799
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr3_+_42107711
|
1.773
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr7_+_28415495
|
1.770
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr9_+_17568952
|
1.727
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr5_+_14493905
|
1.719
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr1_-_16355013
|
1.706
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr22_-_42589543
|
1.696
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr3_+_141136691
|
1.694
|
NM_022131
|
CLSTN2
|
calsyntenin 2
|
|
chr20_+_32610161
|
1.658
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chrX_-_83644054
|
1.656
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chrX_+_110226030
|
1.628
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr5_-_138238891
|
1.609
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr1_+_182040837
|
1.607
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr5_-_138238877
|
1.591
|
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr20_+_19863716
|
1.525
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr14_+_62740878
|
1.501
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr13_-_47885018
|
1.497
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr5_+_66160359
|
1.493
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr22_+_40286957
|
1.488
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr6_+_108593907
|
1.485
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr10_-_95350949
|
1.478
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr14_-_79748273
|
1.470
|
NM_000793
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr10_-_90957028
|
1.454
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr4_+_3341521
|
1.448
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr8_+_134272463
|
1.402
|
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr17_-_46479127
|
1.395
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr8_+_40130143
|
1.387
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr20_-_14266200
|
1.366
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr2_-_134042500
|
1.342
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr4_+_145786622
|
1.338
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr5_+_32747194
|
1.330
|
NM_000908
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr8_-_72436832
|
1.326
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr12_+_12935619
|
1.312
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr2_-_73170290
|
1.307
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr15_-_61460967
|
1.304
|
|
CA12
|
carbonic anhydrase XII
|
|
chr4_-_142273881
|
1.298
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr15_-_61461127
|
1.296
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr4_-_111777651
|
1.286
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr4_-_111777584
|
1.282
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr16_-_4528739
|
1.265
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr2_-_213723298
|
1.262
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr3_-_102194938
|
1.249
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr8_-_72437020
|
1.241
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr1_+_165330710
|
1.235
|
NM_001080426
|
DUSP27
|
dual specificity phosphatase 27 (putative)
|
|
chr19_-_56919032
|
1.233
|
NM_001523
|
HAS1
|
hyaluronan synthase 1
|
|
chr1_+_66570683
|
1.232
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chrX_-_131918905
|
1.228
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr6_+_148705421
|
1.219
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr10_+_63803921
|
1.214
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr14_+_95740768
|
1.213
|
NM_000623
|
BDKRB2
|
bradykinin receptor B2
|
|
chr1_+_66570363
|
1.205
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr13_+_75232797
|
1.200
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr14_+_95792299
|
1.188
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr21_-_42060334
|
1.188
|
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr5_-_139992621
|
1.187
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr5_-_138238953
|
1.168
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr5_-_54317102
|
1.168
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr13_-_47885248
|
1.160
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr1_+_99502655
|
1.158
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr12_-_90029328
|
1.140
|
|
LUM
|
lumican
|
|
chr5_+_118719591
|
1.133
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr4_+_120167264
|
1.126
|
|
SYNPO2
|
synaptopodin 2
|
|
chr4_-_187884721
|
1.111
|
|
|
|
|
chr14_+_100362204
|
1.095
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr21_-_34820916
|
1.088
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr11_-_62213577
|
1.070
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr15_-_81412354
|
1.070
|
NM_004839
NM_199330
NM_199331
NM_199332
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr2_-_31491057
|
1.065
|
|
XDH
|
xanthine dehydrogenase
|
|
chr19_+_18584666
|
1.053
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr4_+_120276386
|
1.044
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr17_-_13445942
|
1.043
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr11_+_108798084
|
1.039
|
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr16_-_71639670
|
1.036
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr2_-_152664056
|
1.034
|
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr13_+_31503436
|
1.031
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr8_-_42184194
|
1.028
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr6_+_42896771
|
1.025
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr8_-_42184350
|
1.023
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr9_-_128924819
|
1.022
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr14_+_100362236
|
1.020
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr22_-_27787793
|
1.018
|
NM_015370
|
C22orf31
|
chromosome 22 open reading frame 31
|
|
chr11_+_108798047
|
1.015
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr15_-_81037554
|
1.011
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr7_-_45095017
|
1.011
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr4_-_156517423
|
1.006
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr2_+_5750229
|
1.006
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr2_-_31491102
|
1.003
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr11_-_8849485
|
1.000
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr6_-_46566969
|
0.999
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr5_+_159276317
|
0.999
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr4_-_156517394
|
0.998
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr5_+_118719448
|
0.990
|
NM_014350
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr3_-_8518327
|
0.989
|
|
LOC100288428
|
hypothetical LOC100288428
|
|
chr4_-_96689373
|
0.988
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr1_-_177106702
|
0.976
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr21_-_42060266
|
0.972
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chrX_+_102471769
|
0.970
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr5_-_94442941
|
0.963
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr8_+_99145925
|
0.962
|
NM_001170806
NM_173549
|
C8orf47
|
chromosome 8 open reading frame 47
|
|
chr4_-_54119159
|
0.947
|
NM_032622
|
LNX1
|
ligand of numb-protein X 1
|
|
chr5_-_111119846
|
0.944
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_85300580
|
0.938
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr12_+_55201854
|
0.937
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr6_+_136214495
|
0.935
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr4_-_54119123
|
0.922
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr10_+_123913094
|
0.920
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr5_-_94443300
|
0.919
|
NM_001002796
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr4_-_54119102
|
0.911
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr20_+_10147455
|
0.908
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr2_-_224175339
|
0.906
|
|
SCG2
|
secretogranin II
|
|
chr8_-_67253234
|
0.902
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr4_+_113958652
|
0.897
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr5_-_147266257
|
0.890
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr12_+_12935759
|
0.887
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr9_-_128924782
|
0.886
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr20_-_33489383
|
0.879
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr12_+_79625538
|
0.879
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr3_+_89239494
|
0.877
|
|
EPHA3
|
EPH receptor A3
|
|
chr4_-_44145580
|
0.874
|
NM_198353
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr4_-_156517544
|
0.873
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chr12_-_90029672
|
0.871
|
NM_002345
|
LUM
|
lumican
|
|
chr4_-_24641404
|
0.864
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr3_+_89239363
|
0.863
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr17_-_19592178
|
0.863
|
NM_000691
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr11_-_8788765
|
0.858
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_-_152663748
|
0.851
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr22_+_39583001
|
0.847
|
NM_022098
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative
|
|
chr6_+_148705646
|
0.838
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr10_-_99780336
|
0.832
|
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr1_+_29435534
|
0.828
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr5_-_139993469
|
0.825
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr11_+_12652427
|
0.817
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr2_-_224611568
|
0.816
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr2_-_188021117
|
0.810
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr2_+_206847658
|
0.809
|
NM_020923
|
ZDBF2
|
zinc finger, DBF-type containing 2
|
|
chr2_-_224175293
|
0.803
|
|
SCG2
|
secretogranin II
|
|
chr1_+_99502435
|
0.795
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr5_-_127901376
|
0.791
|
|
FBN2
|
fibrillin 2
|
|
chr12_-_7935010
|
0.787
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr3_+_151608673
|
0.783
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr17_+_36515166
|
0.776
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr11_+_61204392
|
0.774
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr11_-_62213946
|
0.773
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr8_-_95343716
|
0.771
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr4_-_153493133
|
0.762
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr6_-_46401174
|
0.761
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr2_-_142605010
|
0.754
|
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr11_+_24475091
|
0.751
|
NM_001009909
|
LUZP2
|
leucine zipper protein 2
|
|
chr3_-_56784634
|
0.751
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr9_-_128924706
|
0.747
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr4_-_153820535
|
0.744
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr18_-_35634247
|
0.741
|
|
LOC647946
|
hypothetical LOC647946
|
|
chr8_-_95343696
|
0.737
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr5_+_36642213
|
0.735
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_+_141136896
|
0.730
|
|
CLSTN2
|
calsyntenin 2
|
|
chr15_+_66658626
|
0.725
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr12_+_70120028
|
0.725
|
NM_003667
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr5_+_54491702
|
0.722
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chrX_-_13866565
|
0.714
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr10_-_102663070
|
0.707
|
|
|
|
|
chr11_-_35397675
|
0.707
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr17_+_12510030
|
0.702
|
|
MYOCD
|
myocardin
|
|
chr2_-_206790871
|
0.698
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr2_-_238861830
|
0.688
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr11_-_85107569
|
0.684
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_-_64813459
|
0.682
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr11_-_85108030
|
0.680
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr8_+_94999167
|
0.676
|
NM_001161778
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr5_+_36642424
|
0.671
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_-_159872960
|
0.656
|
NM_020169
|
LXN
|
latexin
|
|
chr3_-_12561962
|
0.656
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr2_+_219779737
|
0.656
|
NM_138802
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr3_-_126257425
|
0.654
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr16_-_4528380
|
0.653
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr2_+_37425240
|
0.653
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr6_-_46401363
|
0.641
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr9_-_32542573
|
0.639
|
NM_001195622
NM_005802
|
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase
|
|
chr6_+_148705817
|
0.634
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr9_-_36390212
|
0.628
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chr4_+_54790190
|
0.624
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr4_+_158361185
|
0.623
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr5_+_73145098
|
0.623
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr1_-_42157077
|
0.620
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|