|
chr20_+_32610161
|
1.274
|
NM_032514
|
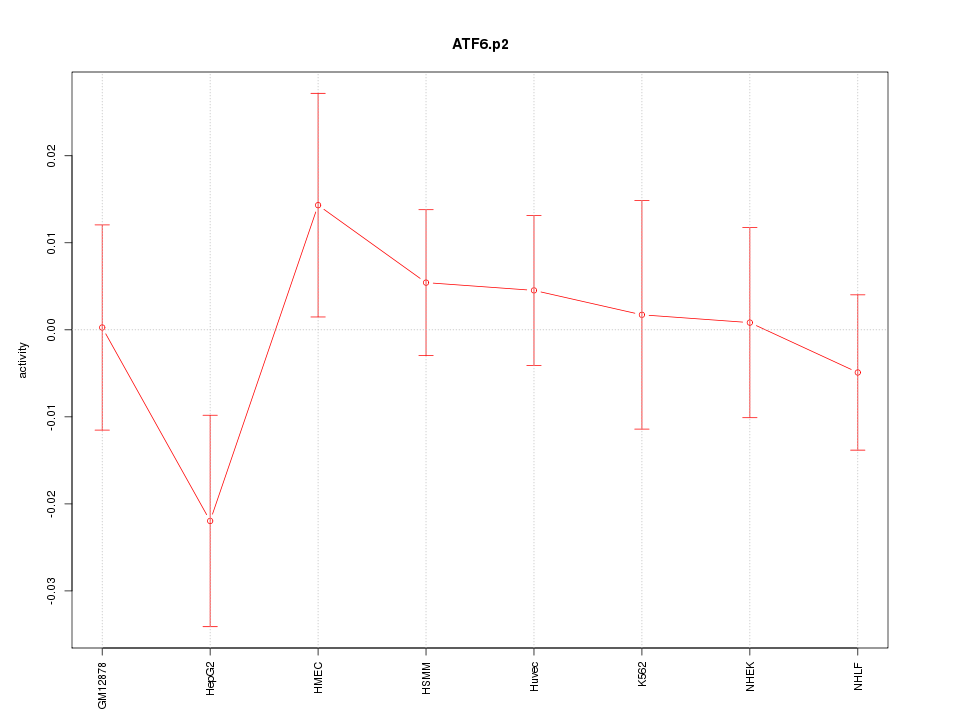
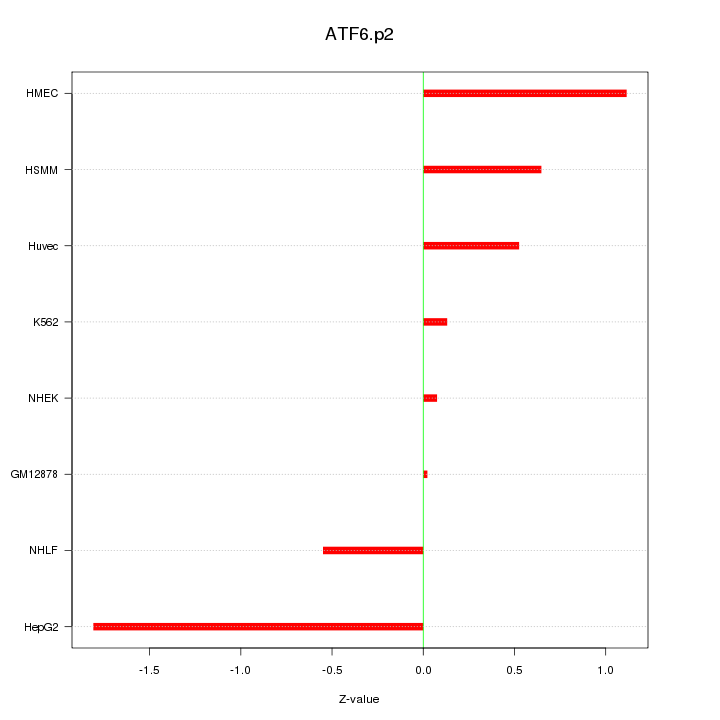
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr1_+_231816372
|
0.991
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr19_+_54723044
|
0.985
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr7_+_100302885
|
0.962
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr1_+_231816540
|
0.890
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr19_-_51608665
|
0.874
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr10_-_7491204
|
0.852
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chrX_-_100801472
|
0.832
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr1_+_13899272
|
0.762
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr8_+_145718702
|
0.737
|
|
|
|
|
chr19_-_45888259
|
0.697
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr8_+_22479115
|
0.666
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr9_-_89779413
|
0.665
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr17_-_44058612
|
0.651
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chrX_+_100692071
|
0.649
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chrX_+_102749489
|
0.646
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr4_-_111763655
|
0.627
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr7_+_27191534
|
0.615
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr11_-_62233600
|
0.614
|
NM_001130702
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr1_+_84539876
|
0.614
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chrX_-_34585287
|
0.608
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr17_-_44043361
|
0.606
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr20_+_19863716
|
0.589
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr7_-_19123765
|
0.580
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr11_-_62233552
|
0.576
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chrX_-_55532355
|
0.569
|
NM_201286
|
USP51
|
ubiquitin specific peptidase 51
|
|
chr12_+_52665196
|
0.568
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr4_-_111763356
|
0.558
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr7_-_19123787
|
0.557
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr17_-_44043278
|
0.552
|
|
HOXB7
|
homeobox B7
|
|
chr1_+_110494654
|
0.537
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr14_-_22458134
|
0.527
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr22_-_42589543
|
0.520
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr14_-_22458199
|
0.509
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr15_-_66511488
|
0.505
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr1_-_50661716
|
0.503
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr5_-_138238891
|
0.502
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr11_-_118739838
|
0.496
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr7_-_35260213
|
0.495
|
NM_001077653
NM_001166220
|
TBX20
|
T-box 20
|
|
chr9_+_35528890
|
0.495
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr17_-_44042924
|
0.490
|
|
HOXB7
|
homeobox B7
|
|
chrX_+_122145776
|
0.489
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr19_-_7459867
|
0.489
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr8_-_9046460
|
0.475
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr9_-_89779180
|
0.473
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr18_+_54682261
|
0.473
|
|
ZNF532
|
zinc finger protein 532
|
|
chr14_-_22458220
|
0.473
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr10_-_113933457
|
0.469
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr11_-_118740069
|
0.468
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_-_35503720
|
0.463
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr1_+_154297588
|
0.461
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr2_+_176695658
|
0.458
|
NM_014213
|
HOXD9
|
homeobox D9
|
|
chr3_-_181237210
|
0.448
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr5_-_138238953
|
0.447
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr5_-_1935764
|
0.443
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr2_-_133144094
|
0.441
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr7_+_116447499
|
0.431
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr9_+_35528628
|
0.423
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr10_-_128067062
|
0.421
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr2_+_219453487
|
0.414
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr4_+_91267706
|
0.409
|
NM_001145065
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr9_-_78710698
|
0.408
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr7_+_100111712
|
0.407
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr7_+_128257698
|
0.404
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr7_+_100041143
|
0.399
|
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr12_+_52634950
|
0.399
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr19_-_52710129
|
0.396
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr7_+_97199206
|
0.395
|
NM_003182
NM_013996
NM_013997
NM_013998
|
TAC1
|
tachykinin, precursor 1
|
|
chr22_+_17273735
|
0.390
|
NM_005675
|
DGCR6
|
DiGeorge syndrome critical region gene 6
|
|
chr10_-_14090459
|
0.390
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_-_202595666
|
0.389
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr3_-_69517913
|
0.387
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr16_-_60627536
|
0.386
|
NM_001796
|
CDH8
|
cadherin 8, type 2
|
|
chr16_+_80370426
|
0.386
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr11_+_113435640
|
0.381
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr16_+_80370400
|
0.375
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr19_-_45016587
|
0.372
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr11_+_108798047
|
0.362
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr11_-_31796061
|
0.360
|
NM_001127612
|
PAX6
|
paired box 6
|
|
chr8_+_54015020
|
0.360
|
NM_005285
|
NPBWR1
|
neuropeptides B/W receptor 1
|
|
chr5_+_170221584
|
0.359
|
NM_022897
|
RANBP17
|
RAN binding protein 17
|
|
chr16_-_80602546
|
0.359
|
NM_145168
|
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr8_-_75396016
|
0.358
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr8_-_24869945
|
0.356
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr22_+_40724674
|
0.354
|
NM_152613
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr10_-_128067052
|
0.354
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr9_+_86474393
|
0.353
|
NM_001018065
NM_001018066
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr7_+_154943466
|
0.350
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr9_+_123501186
|
0.348
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr13_-_94162249
|
0.348
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr10_-_128067001
|
0.346
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr9_+_86474500
|
0.343
|
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr22_-_35502059
|
0.342
|
NM_001177701
NM_006860
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr19_-_62912347
|
0.342
|
NM_001085384
|
ZNF154
|
zinc finger protein 154
|
|
chr2_-_192767494
|
0.341
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr9_+_86474445
|
0.341
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr5_+_159276317
|
0.337
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr9_+_86474457
|
0.337
|
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr9_+_86474553
|
0.334
|
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr8_+_106400322
|
0.332
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr11_+_108798084
|
0.327
|
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr17_-_44010543
|
0.327
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chrX_+_21302317
|
0.325
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr10_-_118022965
|
0.324
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr7_-_27191287
|
0.323
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr4_-_164473107
|
0.323
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr5_+_139924595
|
0.320
|
NM_080670
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr1_-_181654027
|
0.317
|
|
|
|
|
chr21_+_37367432
|
0.316
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr7_+_23686268
|
0.315
|
NM_001127364
NM_001127365
NM_199136
|
C7orf46
|
chromosome 7 open reading frame 46
|
|
chr21_-_43671355
|
0.314
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr11_+_113435506
|
0.313
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr6_-_75972473
|
0.312
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr3_+_50259329
|
0.307
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr19_-_52709975
|
0.307
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr3_-_73756668
|
0.306
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr17_-_72045218
|
0.306
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr3_-_129023850
|
0.297
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr17_-_47591123
|
0.291
|
NM_020178
|
CA10
|
carbonic anhydrase X
|
|
chrX_+_37315934
|
0.290
|
|
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial)
|
|
chr15_-_70399551
|
0.284
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr1_+_148521852
|
0.283
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr1_+_4614528
|
0.283
|
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr7_-_5428043
|
0.282
|
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr11_-_65138217
|
0.282
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr11_-_57173823
|
0.281
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chrX_+_133952633
|
0.280
|
NM_001163438
|
LOC644538
|
hypothetical protein LOC644538
|
|
chr19_+_54149941
|
0.280
|
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_155053124
|
0.279
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr19_-_52710089
|
0.279
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr10_-_128066935
|
0.278
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr17_-_47590975
|
0.276
|
|
CA10
|
carbonic anhydrase X
|
|
chr19_+_43502360
|
0.275
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr19_+_43502380
|
0.275
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr7_+_100111689
|
0.274
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr12_-_14612057
|
0.272
|
NM_024829
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr3_-_69518119
|
0.272
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr13_+_24940965
|
0.271
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr19_+_43502591
|
0.271
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr15_+_39739864
|
0.270
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr19_+_43502301
|
0.268
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr12_+_64869181
|
0.267
|
NM_001142523
NM_007199
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr5_-_37875538
|
0.265
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr3_-_12775718
|
0.265
|
NM_018306
|
TMEM40
|
transmembrane protein 40
|
|
chr9_+_130749791
|
0.263
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr19_-_12891074
|
0.262
|
NM_001105578
|
SYCE2
|
synaptonemal complex central element protein 2
|
|
chr11_+_125279481
|
0.262
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr1_+_221633337
|
0.260
|
NM_152610
|
C1orf65
|
chromosome 1 open reading frame 65
|
|
chr5_+_170221441
|
0.259
|
|
RANBP17
|
RAN binding protein 17
|
|
chr1_+_220857774
|
0.259
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr7_+_28415495
|
0.259
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr12_+_20413445
|
0.258
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr12_-_14611820
|
0.258
|
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr15_-_70399523
|
0.257
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr19_+_54149843
|
0.254
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr19_+_54149963
|
0.252
|
|
BAX
|
BCL2-associated X protein
|
|
chr12_+_64869271
|
0.250
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr13_-_35818396
|
0.249
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr6_-_10527772
|
0.248
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr3_-_24511456
|
0.248
|
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr4_-_2905448
|
0.248
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr4_-_100069303
|
0.246
|
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chrX_+_37315740
|
0.246
|
NM_001170331
NM_198511
|
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial)
|
|
chr8_-_11096257
|
0.246
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr11_+_68208554
|
0.245
|
NM_015973
|
GAL
|
galanin prepropeptide
|
|
chr14_+_100362204
|
0.243
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr11_+_13646806
|
0.241
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr1_-_43509187
|
0.239
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr7_-_8268206
|
0.238
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr15_+_39739933
|
0.237
|
|
MGA
|
MAX gene associated
|
|
chr20_-_10148137
|
0.237
|
|
LOC100131208
|
similar to hCG2045825
|
|
chr13_+_24940917
|
0.237
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr10_-_62373828
|
0.237
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chrX_+_44617346
|
0.237
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr3_+_62279707
|
0.235
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr11_+_68208583
|
0.233
|
|
GAL
|
galanin prepropeptide
|
|
chr4_-_104860277
|
0.233
|
NM_001059
|
TACR3
|
tachykinin receptor 3
|
|
chr1_+_11718715
|
0.227
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr12_+_4788602
|
0.226
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr11_+_13646727
|
0.225
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr3_-_24511180
|
0.224
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr11_+_13646822
|
0.223
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr12_-_47604930
|
0.222
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr2_-_96174865
|
0.222
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr2_-_1727292
|
0.222
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr3_+_114733907
|
0.222
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr10_-_32257693
|
0.222
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chrX_-_47394937
|
0.221
|
NM_001114123
NM_005229
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr14_-_66948523
|
0.221
|
NM_016445
|
PLEK2
|
pleckstrin 2
|
|
chr7_-_8268292
|
0.220
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr9_-_103289154
|
0.219
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr11_-_65138323
|
0.218
|
|
|
|
|
chr1_-_226357552
|
0.217
|
NM_024319
|
C1orf35
|
chromosome 1 open reading frame 35
|
|
chr10_+_70517823
|
0.217
|
NM_002727
|
SRGN
|
serglycin
|
|
chr3_+_148609841
|
0.215
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chrX_-_139414890
|
0.215
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr12_-_112394259
|
0.215
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr6_-_75972286
|
0.215
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr4_+_43185
|
0.213
|
NM_182524
|
ZNF595
ZNF718
|
zinc finger protein 595
zinc finger protein 718
|
|
chr2_-_96174502
|
0.212
|
|
DUSP2
|
dual specificity phosphatase 2
|