|
chr18_+_75825563
|
2.632
|
NM_001136180
|
HSBP1L1
|
heat shock factor binding protein 1-like 1
|
|
chr20_+_54637714
|
2.212
|
NM_003222
|
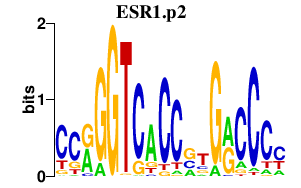
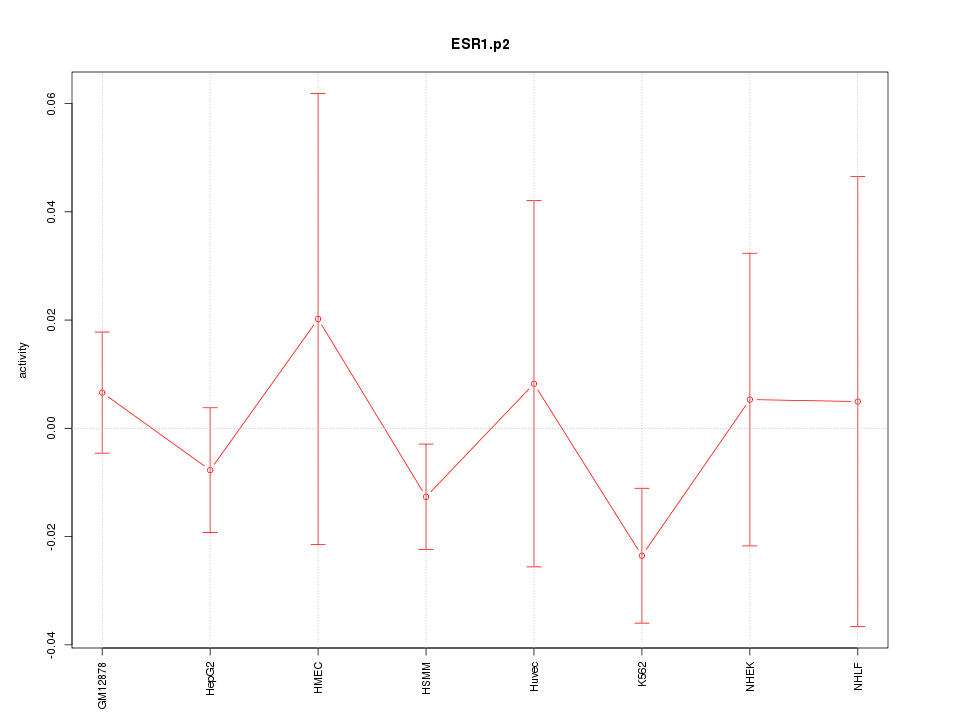
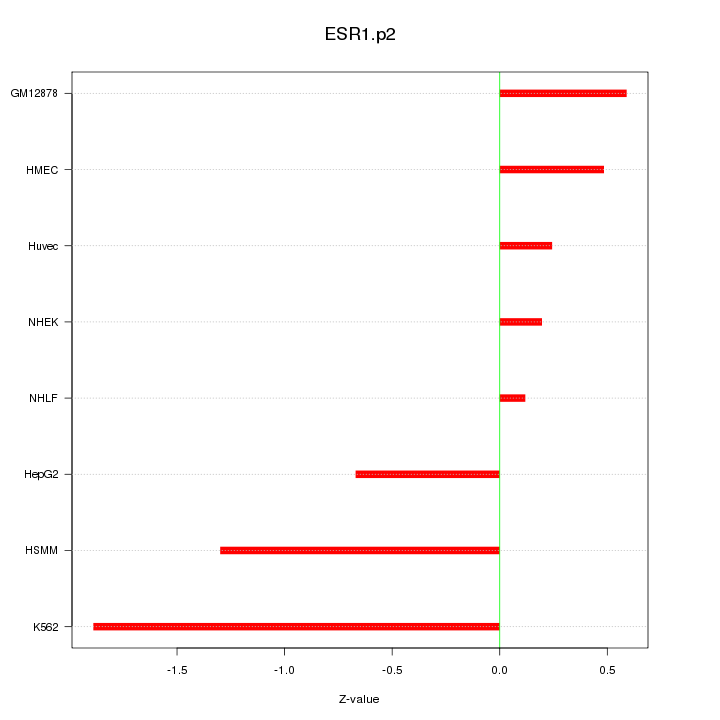
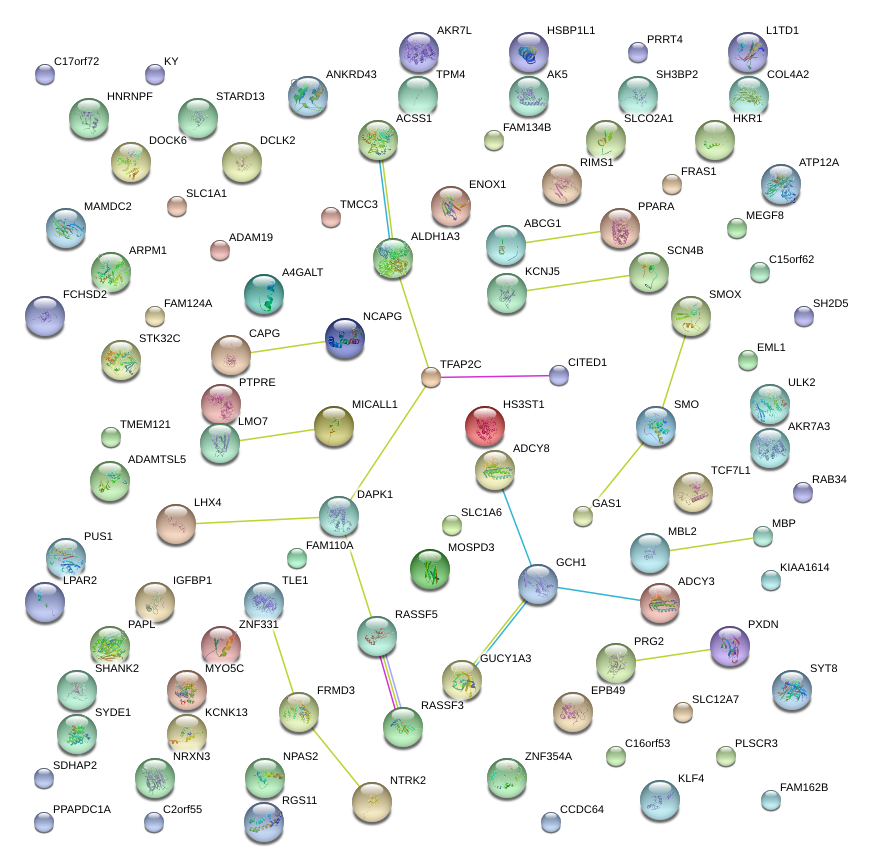
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr17_-_7238546
|
1.905
|
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr17_-_24068759
|
1.793
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr1_+_62433061
|
1.578
|
NM_001164835
NM_019079
|
L1TD1
|
LINE-1 type transposase domain containing 1
|
|
chr19_-_19599973
|
1.501
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr1_+_178466064
|
1.398
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chr16_+_88305743
|
1.388
|
|
LOC100128881
|
hypothetical LOC100128881
|
|
chr22_+_36632242
|
1.368
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr1_+_179148935
|
1.349
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr3_-_170970296
|
1.332
|
NM_032487
|
ARPM1
|
actin related protein M1
|
|
chr14_+_78815406
|
1.332
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr19_-_14951349
|
1.329
|
|
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6
|
|
chr22_-_41447214
|
1.308
|
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr13_+_24152548
|
1.296
|
NM_001185085
NM_001676
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide
|
|
chr18_-_72973698
|
1.288
|
|
MBP
|
myelin basic protein
|
|
chr1_-_29322993
|
1.285
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr10_+_129595289
|
1.284
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr9_-_109290107
|
1.276
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr9_-_85342868
|
1.266
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr16_+_29734785
|
1.256
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr11_+_128266460
|
1.241
|
NM_000890
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5
|
|
chr11_+_1812115
|
1.236
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chrX_-_71442488
|
1.188
|
NM_001144885
NM_001144887
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1
|
|
chr18_-_72973750
|
1.187
|
NM_001025100
NM_001025101
|
MBP
|
myelin basic protein
|
|
chr19_+_58733144
|
1.185
|
NM_001079906
|
ZNF331
|
zinc finger protein 331
|
|
chr21_+_42512999
|
1.176
|
NM_207174
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr14_-_54438913
|
1.165
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr19_+_42517337
|
1.164
|
NM_181786
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member
|
|
chr1_+_77520315
|
1.150
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr4_-_11039600
|
1.130
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr3_-_135852514
|
1.127
|
NM_178554
|
KY
|
kyphoscoliosis peptidase
|
|
chr5_-_16670085
|
1.122
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr12_-_93568336
|
1.115
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr19_+_58750305
|
1.111
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr11_-_117528744
|
1.107
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr7_+_128615948
|
1.107
|
NM_005631
|
SMO
|
smoothened homolog (Drosophila)
|
|
chr10_-_133971136
|
1.091
|
NM_173575
|
STK32C
|
serine/threonine kinase 32C
|
|
chr4_+_151218862
|
1.085
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr13_-_32822759
|
1.080
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_+_75108450
|
1.076
|
|
LMO7
|
LIM domain 7
|
|
chr13_+_109757593
|
1.072
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr15_+_38849450
|
1.068
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr13_-_43259032
|
1.062
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr6_+_30002084
|
1.057
|
|
|
|
|
chr19_+_15079141
|
1.056
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr9_+_45331898
|
1.012
|
|
LOC100132167
|
hypothetical LOC100132167
|
|
chr6_+_72949092
|
1.002
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_20931288
|
0.982
|
|
SH2D5
|
SH2 domain containing 5
|
|
chr3_-_135231417
|
0.972
|
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr12_-_123568781
|
0.970
|
|
|
|
|
chr16_-_265868
|
0.968
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr9_+_86473273
|
0.963
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr19_+_47521600
|
0.957
|
NM_001410
|
MEGF8
|
multiple EGF-like-domains 8
|
|
chr17_+_59429442
|
0.956
|
NM_001164257
NM_001191029
NM_001191030
NM_001191031
|
C17orf72
|
chromosome 17 open reading frame 72
|
|
chr1_+_77520874
|
0.948
|
NM_012093
|
AK5
|
adenylate kinase 5
|
|
chr4_+_156807311
|
0.940
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr1_-_19487790
|
0.935
|
NM_012067
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase)
|
|
chr2_-_98919109
|
0.931
|
NM_207362
|
C2orf55
|
chromosome 2 open reading frame 55
|
|
chr4_+_79197747
|
0.916
|
NM_001166133
NM_025074
|
FRAS1
|
Fraser syndrome 1
|
|
chr11_-_72530605
|
0.915
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr2_-_85494664
|
0.910
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr10_+_122206455
|
0.910
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr15_-_50375143
|
0.909
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr20_-_24986501
|
0.902
|
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr21_+_42512271
|
0.900
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr15_+_99237555
|
0.898
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr11_-_70641267
|
0.885
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr9_-_88751923
|
0.880
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr9_-_83493415
|
0.873
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr7_+_45894480
|
0.867
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr2_-_85494639
|
0.865
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr19_-_11234117
|
0.863
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr22_-_41446740
|
0.861
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr5_-_156935283
|
0.859
|
|
|
|
|
chr12_+_130979696
|
0.858
|
NM_001002020
|
PUS1
|
pseudouridylate synthase 1
|
|
chr19_+_16039170
|
0.847
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr19_+_44266784
|
0.845
|
NM_001004318
|
PAPL
|
iron/zinc purple acid phosphatase-like protein
|
|
chr7_-_127788893
|
0.844
|
NM_001114726
NM_001174164
|
PRRT4
|
proline-rich transmembrane protein 4
|
|
chr14_+_99273751
|
0.844
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr15_+_99237527
|
0.838
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr9_+_71848555
|
0.837
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr14_+_89597860
|
0.833
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr19_-_1463997
|
0.832
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr17_-_19711580
|
0.830
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr4_+_156808261
|
0.827
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr2_-_24995402
|
0.826
|
NM_004036
|
ADCY3
|
adenylate cyclase 3
|
|
chr9_+_4480193
|
0.825
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr2_+_85213884
|
0.823
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr2_+_100802923
|
0.819
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr20_+_773283
|
0.808
|
NM_031424
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr4_+_2790338
|
0.807
|
NM_003023
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr12_+_118911859
|
0.804
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr14_+_105063975
|
0.803
|
NM_025268
|
TMEM121
|
transmembrane protein 121
|
|
chr12_+_130979729
|
0.800
|
NM_001002019
NM_025215
|
PUS1
|
pseudouridylate synthase 1
|
|
chr3_-_135231518
|
0.794
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr5_+_132176921
|
0.784
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr5_-_178090203
|
0.779
|
NM_005649
|
ZNF354A
|
zinc finger protein 354A
|
|
chr5_-_1165106
|
0.771
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr22_+_44925162
|
0.767
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr3_-_197201497
|
0.766
|
|
SDHAP1
SDHAP2
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 1
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2
|
|
chr13_+_50694364
|
0.764
|
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr5_-_156935317
|
0.763
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr7_+_100047660
|
0.761
|
NM_001040097
|
MOSPD3
|
motile sperm domain containing 3
|
|
chr9_+_89302574
|
0.760
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr6_-_117193505
|
0.759
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr8_+_21980315
|
0.757
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr1_+_204747483
|
0.756
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr1_+_204747486
|
0.752
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr10_-_43224604
|
0.752
|
NM_004966
NM_001098204
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr10_-_43224312
|
0.749
|
NM_001098205
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr7_+_6622051
|
0.746
|
NM_017560
|
ZNF853
|
zinc finger protein 853
|
|
chr5_-_37285160
|
0.740
|
NM_023073
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr19_-_41191423
|
0.736
|
NM_001039876
|
C19orf46
|
chromosome 19 open reading frame 46
|
|
chr1_+_77520190
|
0.736
|
|
AK5
|
adenylate kinase 5
|
|
chr3_+_195889845
|
0.733
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr22_+_38720881
|
0.732
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr20_-_3696344
|
0.728
|
NM_001039140
|
C20orf27
|
chromosome 20 open reading frame 27
|
|
chr4_+_37569093
|
0.726
|
NM_015173
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr1_-_19473021
|
0.726
|
NM_001145289
NM_201252
|
AKR7L
|
aldo-keto reductase family 7-like
|
|
chr21_-_26867138
|
0.723
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr1_+_170077234
|
0.722
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr13_-_27572646
|
0.721
|
NM_004119
|
FLT3
|
fms-related tyrosine kinase 3
|
|
chr19_+_61711008
|
0.721
|
NM_020813
|
ZNF471
|
zinc finger protein 471
|
|
chr11_+_128067598
|
0.720
|
NM_001167681
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr10_-_105410840
|
0.715
|
|
|
|
|
chr1_+_208178160
|
0.715
|
NM_001146261
NM_001146262
NM_001146264
NM_153262
|
SYT14
|
synaptotagmin XIV
|
|
chr22_-_49413326
|
0.714
|
NM_000487
NM_001085425
NM_001085426
NM_001085427
NM_001085428
|
ARSA
|
arylsulfatase A
|
|
chr11_-_63138516
|
0.714
|
NM_001128203
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr16_+_81218142
|
0.710
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr16_+_29734990
|
0.708
|
NM_024516
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr5_-_178983241
|
0.703
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr11_-_605998
|
0.701
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr4_+_1842833
|
0.701
|
NM_001042424
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr5_+_110587546
|
0.699
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr1_+_15446354
|
0.698
|
NM_052929
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1
|
|
chr7_-_28964186
|
0.698
|
|
|
|
|
chr5_-_178983281
|
0.691
|
NM_005520
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr4_+_184257337
|
0.690
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr12_+_3470685
|
0.690
|
NM_019854
|
PRMT8
|
protein arginine methyltransferase 8
|
|
chr19_-_10091534
|
0.685
|
NM_003755
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G
|
|
chr1_+_208178198
|
0.685
|
|
SYT14
|
synaptotagmin XIV
|
|
chr13_-_25523164
|
0.685
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr10_-_49483002
|
0.684
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr2_+_119697859
|
0.684
|
|
STEAP3
|
STEAP family member 3
|
|
chr19_+_41177882
|
0.682
|
NM_001042631
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1
|
|
chr11_-_605920
|
0.682
|
|
IRF7
|
interferon regulatory factor 7
|
|
chr18_-_45975163
|
0.676
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr14_+_53933422
|
0.672
|
NM_001130851
NM_005192
|
CDKN3
|
cyclin-dependent kinase inhibitor 3
|
|
chr11_-_63138416
|
0.671
|
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr21_+_32167175
|
0.670
|
NM_014586
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr11_-_63138483
|
0.670
|
NM_007069
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr14_+_73073570
|
0.668
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr11_-_900793
|
0.660
|
|
CHID1
|
chitinase domain containing 1
|
|
chr16_-_674357
|
0.660
|
NM_001005920
|
JMJD8
|
jumonji domain containing 8
|
|
chr2_-_53940571
|
0.659
|
NM_001164165
NM_006794
|
GPR75-ASB3
GPR75
|
GPR75-ASB3 read-through transcript
G protein-coupled receptor 75
|
|
chr11_-_1549719
|
0.658
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr11_-_63138061
|
0.657
|
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr13_+_43351968
|
0.652
|
NM_153218
|
C13orf31
|
chromosome 13 open reading frame 31
|
|
chr2_-_42574587
|
0.650
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr21_-_37274797
|
0.648
|
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr11_+_63883192
|
0.648
|
NM_001006944
NM_003942
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4
|
|
chr5_+_178219519
|
0.646
|
NM_058230
|
ZNF354B
|
zinc finger protein 354B
|
|
chr6_+_107917965
|
0.645
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr16_+_81218074
|
0.644
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr2_+_149894744
|
0.644
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr15_-_88034857
|
0.634
|
NM_003847
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha
|
|
chr11_-_75599433
|
0.633
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr1_+_61980679
|
0.631
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr6_+_74029599
|
0.630
|
|
|
|
|
chr17_-_30724732
|
0.628
|
NM_001104587
NM_001104588
NM_001104589
NM_001104590
NM_152270
|
SLFN11
|
schlafen family member 11
|
|
chr19_-_1464187
|
0.628
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr10_+_102748062
|
0.628
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr2_-_198358253
|
0.625
|
NM_033030
|
BOLL
|
bol, boule-like (Drosophila)
|
|
chr10_-_126839546
|
0.622
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr16_-_1600654
|
0.618
|
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas)
|
|
chr21_-_44485269
|
0.616
|
|
ICOSLG
|
inducible T-cell co-stimulator ligand
|
|
chr3_+_48239865
|
0.613
|
NM_004345
|
CAMP
|
cathelicidin antimicrobial peptide
|
|
chr4_-_89299034
|
0.609
|
NM_004827
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2
|
|
chr1_+_154390939
|
0.609
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr20_+_41976831
|
0.607
|
NM_001098797
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr1_+_15352800
|
0.606
|
NM_001136218
NM_018022
|
TMEM51
|
transmembrane protein 51
|
|
chr7_+_100141609
|
0.606
|
NM_005837
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr14_+_95575319
|
0.604
|
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr6_+_158877455
|
0.599
|
NM_020823
|
TMEM181
|
transmembrane protein 181
|
|
chr12_+_52689062
|
0.598
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr9_+_137532294
|
0.598
|
NM_016034
|
MRPS2
|
mitochondrial ribosomal protein S2
|
|
chr11_+_63463009
|
0.596
|
NM_024771
|
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae)
|
|
chr15_+_38923914
|
0.595
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr7_+_2638156
|
0.595
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr6_+_30082331
|
0.595
|
|
HLA-J
|
major histocompatibility complex, class I, J (pseudogene)
|
|
chr18_-_42590989
|
0.594
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr17_-_45562112
|
0.593
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr14_+_64076916
|
0.593
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr20_-_60415729
|
0.589
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr5_-_131375148
|
0.589
|
NM_001009185
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr9_+_111850698
|
0.586
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr21_-_42303517
|
0.586
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr19_+_1442000
|
0.585
|
NM_138393
|
REEP6
|
receptor accessory protein 6
|
|
chr22_-_45024820
|
0.585
|
NM_207327
|
C22orf40
|
chromosome 22 open reading frame 40
|