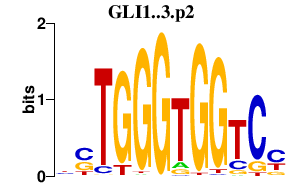
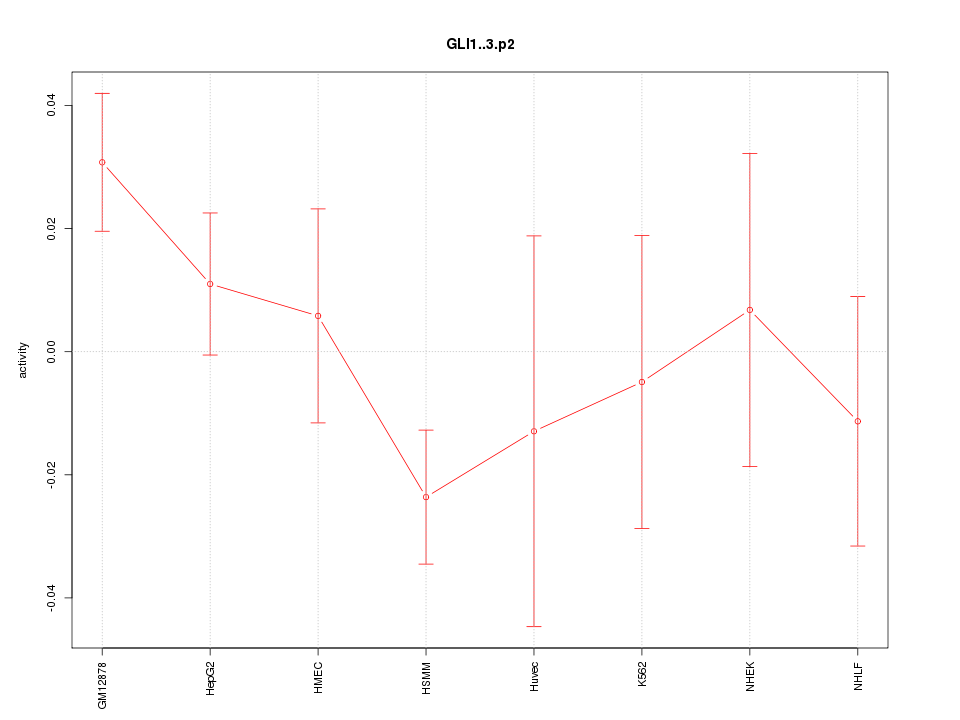
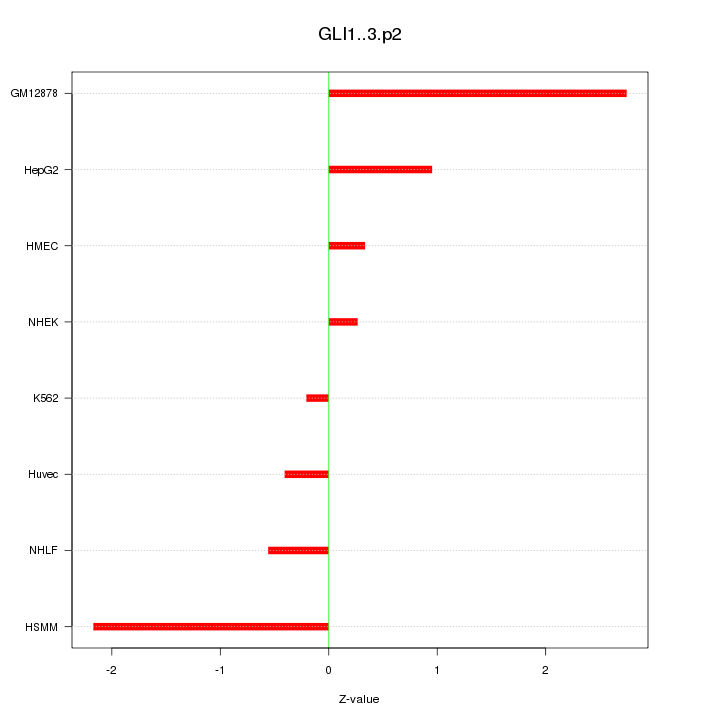
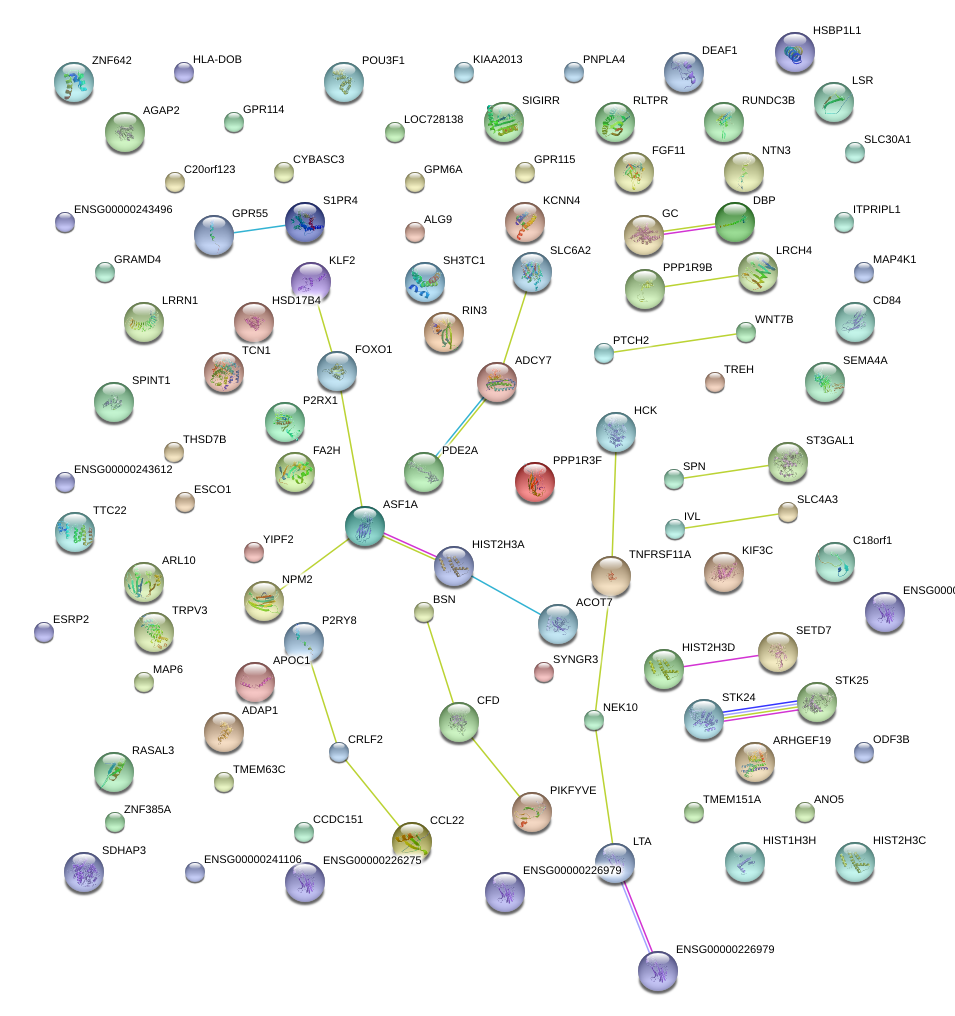
|
chr6_-_32892705
|
2.775
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr11_-_60880799
|
2.118
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr11_-_72057755
|
1.966
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr18_+_75825563
|
1.797
|
NM_001136180
|
HSBP1L1
|
heat shock factor binding protein 1-like 1
|
|
chr16_+_48865521
|
1.733
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr1_+_154389945
|
1.569
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr5_+_175725050
|
1.531
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr2_-_231498184
|
1.490
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr17_-_3407780
|
1.448
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr12_-_56422120
|
1.437
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr16_-_66827449
|
1.430
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr1_-_16411690
|
1.352
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr19_+_38485583
|
1.338
|
|
LOC80054
|
hypothetical LOC80054
|
|
chr2_-_26058831
|
1.316
|
|
KIF3C
|
kinesin family member 3C
|
|
chr1_+_151147646
|
1.291
|
NM_005547
|
IVL
|
involucrin
|
|
chrX_-_1615999
|
1.284
|
NM_178129
|
P2RY8
CRLF2
|
purinergic receptor P2Y, G-protein coupled, 8
cytokine receptor-like factor 2
|
|
chr6_+_44076281
|
1.254
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr18_-_17434811
|
1.241
|
|
ESCO1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae)
|
|
chr1_-_55039458
|
1.171
|
NM_001114108
NM_017904
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr1_-_38285033
|
1.165
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr19_+_3129735
|
1.160
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr19_-_15436333
|
1.153
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr16_-_3049336
|
1.152
|
|
|
|
|
chr17_-_3408038
|
1.139
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr7_+_87095640
|
1.124
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr8_+_21937533
|
1.103
|
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr17_-_37181626
|
1.096
|
|
|
|
|
chr19_-_48976852
|
1.096
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr22_-_49317408
|
1.076
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr16_+_66236512
|
1.041
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr1_-_11909042
|
1.034
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr11_+_9736408
|
1.031
|
|
LOC283104
|
hypothetical LOC283104
|
|
chr19_-_48977247
|
1.029
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_+_40715465
|
1.017
|
|
ZNF642
|
zinc finger protein 642
|
|
chr19_+_2240996
|
1.008
|
|
|
|
|
chr16_+_66236559
|
1.002
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr3_+_49566901
|
1.000
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr1_-_209818675
|
0.988
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr16_+_56134077
|
0.970
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr3_-_27385856
|
0.964
|
NM_199347
|
NEK10
|
NIMA (never in mitosis gene a)- related kinase 10
|
|
chr17_+_7283412
|
0.961
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr2_+_96355661
|
0.958
|
NM_178495
NM_001163524
|
ITPRIPL1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr16_+_56134037
|
0.951
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr14_-_106330781
|
0.949
|
|
IGHV5-78
|
immunoglobulin heavy variable 5-78 pseudogene
|
|
chr4_-_140696740
|
0.945
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr16_+_1979959
|
0.930
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr19_+_40431678
|
0.923
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chrX_+_49013424
|
0.919
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr2_+_137239583
|
0.905
|
|
THSD7B
|
thrombospondin, type I, domain containing 7B
|
|
chr11_+_65815911
|
0.899
|
|
TMEM151A
|
transmembrane protein 151A
|
|
chr12_+_6915698
|
0.895
|
|
|
|
|
chr5_+_176717553
|
0.893
|
|
|
|
|
chr19_+_16296632
|
0.887
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr19_+_811615
|
0.882
|
|
CFD
|
complement factor D (adipsin)
|
|
chr4_-_176970895
|
0.874
|
|
GPM6A
|
glycoprotein M6A
|
|
chr11_-_404975
|
0.865
|
NM_001135054
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr5_-_1647617
|
0.864
|
|
SDHAP3
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3
|
|
chr2_+_220200530
|
0.861
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr3_+_3816079
|
0.828
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr11_-_118055564
|
0.814
|
NM_007180
|
TREH
|
trehalase (brush-border membrane glycoprotein)
|
|
chr1_-_209818576
|
0.809
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr16_-_73366200
|
0.802
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr6_+_31647854
|
0.800
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr17_-_3766441
|
0.796
|
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1
|
|
chr16_+_29581783
|
0.789
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr22_+_45401321
|
0.787
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr16_+_54247852
|
0.786
|
NM_001043
NM_001172504
|
SLC6A2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2
|
|
chr11_+_65815940
|
0.783
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr19_+_16296734
|
0.780
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chrX_-_7855764
|
0.776
|
NM_001142389
NM_001172672
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr15_+_38923914
|
0.776
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr2_-_242096673
|
0.768
|
NM_006374
|
STK25
|
serine/threonine kinase 25
|
|
chr13_-_40138607
|
0.768
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr1_-_158815850
|
0.753
|
|
CD84
|
CD84 molecule
|
|
chr12_-_53064723
|
0.753
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr18_-_17434655
|
0.752
|
NM_052911
|
ESCO1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae)
|
|
chr11_-_75056761
|
0.749
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr19_+_50109903
|
0.747
|
|
APOC1
|
apolipoprotein C-I
|
|
chr19_-_10900086
|
0.747
|
NM_024029
|
YIPF2
|
Yip1 domain family, member 2
|
|
chr8_-_134653194
|
0.745
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr20_+_61655934
|
0.743
|
|
|
|
|
chr19_-_43800374
|
0.739
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr16_+_55950195
|
0.739
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr6_+_47774247
|
0.735
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr22_-_44751671
|
0.732
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr7_-_960534
|
0.729
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr4_+_8251818
|
0.726
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr1_-_45081190
|
0.725
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr11_+_22171150
|
0.721
|
NM_001142649
NM_213599
|
ANO5
|
anoctamin 5
|
|
chr4_-_176970372
|
0.720
|
|
GPM6A
|
glycoprotein M6A
|
|
chr16_-_73365975
|
0.717
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr14_+_76717835
|
0.715
|
NM_020431
|
TMEM63C
|
transmembrane protein 63C
|
|
chr14_+_92188546
|
0.715
|
|
RIN3
|
Ras and Rab interactor 3
|
|
chr1_-_6343306
|
0.713
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr20_+_30103642
|
0.712
|
NM_001172129
NM_001172130
NM_001172131
NM_001172132
NM_001172133
NM_002110
|
HCK
|
hemopoietic cell kinase
|
|
chr18_+_58143499
|
0.710
|
NM_003839
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator
|
|
chr2_+_19932101
|
0.703
|
|
FLJ12334
|
hypothetical LOC400946
|
|
chr19_-_53832381
|
0.703
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr20_-_44612619
|
0.703
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chr3_+_50272648
|
0.700
|
|
|
|
|
chr18_+_13208777
|
0.698
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr16_+_2461500
|
0.696
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr1_+_148090804
|
0.696
|
NM_021059
NM_001005464
|
HIST2H3C
HIST2H3A
|
histone cluster 2, H3c
histone cluster 2, H3a
|
|
chr3_+_47004457
|
0.693
|
|
NBEAL2
|
neurobeachin-like 2
|
|
chr6_+_3009120
|
0.692
|
|
RIPK1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1
|
|
chr2_-_242096592
|
0.689
|
|
STK25
|
serine/threonine kinase 25
|
|
chr4_-_176970407
|
0.685
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_957269
|
0.680
|
|
DGKQ
|
diacylglycerol kinase, theta 110kDa
|
|
chr5_-_156935283
|
0.677
|
|
|
|
|
chr20_-_42457717
|
0.674
|
|
|
|
|
chr5_-_1165106
|
0.673
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr16_+_30952350
|
0.672
|
|
STX4
|
syntaxin 4
|
|
chr5_+_176717449
|
0.671
|
NM_006480
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr17_-_74231240
|
0.667
|
|
CYTH1
|
cytohesin 1
|
|
chr17_+_4795128
|
0.667
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr17_-_71272811
|
0.666
|
NM_000154
|
GALK1
|
galactokinase 1
|
|
chr7_+_149699185
|
0.659
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr16_-_11588525
|
0.656
|
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr17_+_45008473
|
0.651
|
|
NXPH3
|
neurexophilin 3
|
|
chr1_+_34993234
|
0.641
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr7_+_99809068
|
0.639
|
|
PILRA
|
paired immunoglobin-like type 2 receptor alpha
|
|
chr2_-_6923215
|
0.639
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr11_+_112763722
|
0.639
|
NM_178510
|
ANKK1
|
ankyrin repeat and kinase domain containing 1
|
|
chr1_-_6341553
|
0.638
|
NM_181866
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr17_-_37181636
|
0.637
|
|
JUP
|
junction plakoglobin
|
|
chr16_+_56259657
|
0.635
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr11_+_75157450
|
0.632
|
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr11_-_30564338
|
0.629
|
NM_001145399
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr12_+_110328126
|
0.629
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr17_-_34607257
|
0.624
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr2_+_46623325
|
0.622
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr9_+_130258256
|
0.620
|
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr12_-_6668680
|
0.619
|
|
ZNF384
|
zinc finger protein 384
|
|
chr20_+_61654816
|
0.619
|
NM_024059
|
C20orf195
|
chromosome 20 open reading frame 195
|
|
chr14_-_106352471
|
0.618
|
|
|
|
|
chr7_-_148212336
|
0.616
|
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr19_+_3087026
|
0.614
|
NM_002068
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class)
|
|
chr1_+_153513810
|
0.614
|
NM_020897
|
HCN3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3
|
|
chr17_+_73738985
|
0.613
|
NM_001145529
|
TMEM235
|
transmembrane protein 235
|
|
chr22_-_49317873
|
0.611
|
NM_001014440
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr20_+_31783426
|
0.610
|
NM_032819
|
ZNF341
|
zinc finger protein 341
|
|
chr5_-_156935317
|
0.608
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr19_+_10900502
|
0.605
|
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr12_-_101835026
|
0.602
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr1_+_199259664
|
0.601
|
|
|
|
|
chrX_+_48427103
|
0.599
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr9_-_138757175
|
0.599
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr13_-_24643620
|
0.598
|
|
FAM123A
|
family with sequence similarity 123A
|
|
chr17_-_45617899
|
0.595
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr15_-_76720598
|
0.595
|
NM_000750
|
CHRNB4
|
cholinergic receptor, nicotinic, beta 4
|
|
chr7_+_72883128
|
0.592
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr2_-_85490984
|
0.591
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr4_+_100957003
|
0.590
|
NM_014395
|
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides
|
|
chr4_-_176971175
|
0.589
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr11_-_118405288
|
0.587
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr19_+_63670148
|
0.586
|
NM_014347
|
ZNF324
|
zinc finger protein 324
|
|
chr19_-_11477689
|
0.586
|
NM_138783
|
ZNF653
|
zinc finger protein 653
|
|
chr10_+_8137356
|
0.585
|
|
GATA3
|
GATA binding protein 3
|
|
chr16_-_10184111
|
0.584
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr1_-_39929654
|
0.581
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr7_-_99518294
|
0.579
|
|
ZNF3
|
zinc finger protein 3
|
|
chr11_+_65311142
|
0.579
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr20_-_43974092
|
0.579
|
NM_006227
NM_182676
|
PLTP
|
phospholipid transfer protein
|
|
chr7_-_102044374
|
0.578
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr10_+_85944383
|
0.578
|
NM_001171971
NM_033100
|
CDHR1
|
cadherin-related family member 1
|
|
chr14_-_50631407
|
0.578
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr10_+_71060008
|
0.578
|
NM_145306
|
C10orf35
|
chromosome 10 open reading frame 35
|
|
chr5_+_150206277
|
0.577
|
NM_001145805
|
IRGM
|
immunity-related GTPase family, M
|
|
chr9_+_130258100
|
0.574
|
NM_153432
NM_153435
NM_153439
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr16_+_30952382
|
0.574
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr17_-_17340219
|
0.574
|
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr12_+_45896511
|
0.573
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr10_+_99488223
|
0.572
|
NM_001002261
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr17_-_53920699
|
0.569
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chr12_-_53064565
|
0.569
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr15_+_37660568
|
0.568
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr6_-_133126278
|
0.566
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr3_-_120761092
|
0.566
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr6_+_33280396
|
0.565
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr20_+_44180355
|
0.563
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr2_+_172658453
|
0.559
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr13_-_19665100
|
0.556
|
NM_004004
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr1_-_210939727
|
0.554
|
|
|
|
|
chr7_+_73826244
|
0.553
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr3_-_53055078
|
0.552
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr3_-_159306214
|
0.548
|
|
|
|
|
chr7_-_127833231
|
0.546
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr19_+_482737
|
0.546
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr12_+_6517600
|
0.544
|
|
|
|
|
chr9_+_130258247
|
0.543
|
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr2_-_26058941
|
0.540
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr1_-_111544820
|
0.538
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr20_+_44180296
|
0.538
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr8_+_21938299
|
0.537
|
NM_182795
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr19_-_38485281
|
0.534
|
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chrX_+_106336517
|
0.531
|
NM_001169154
NM_173494
|
CXorf41
|
chromosome X open reading frame 41
|
|
chr13_-_19665036
|
0.531
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr11_-_83071084
|
0.531
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr12_-_101396459
|
0.530
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr18_+_2896178
|
0.528
|
|
EMILIN2
|
elastin microfibril interfacer 2
|