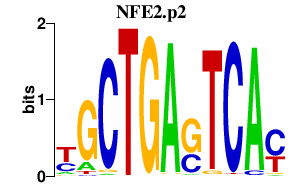
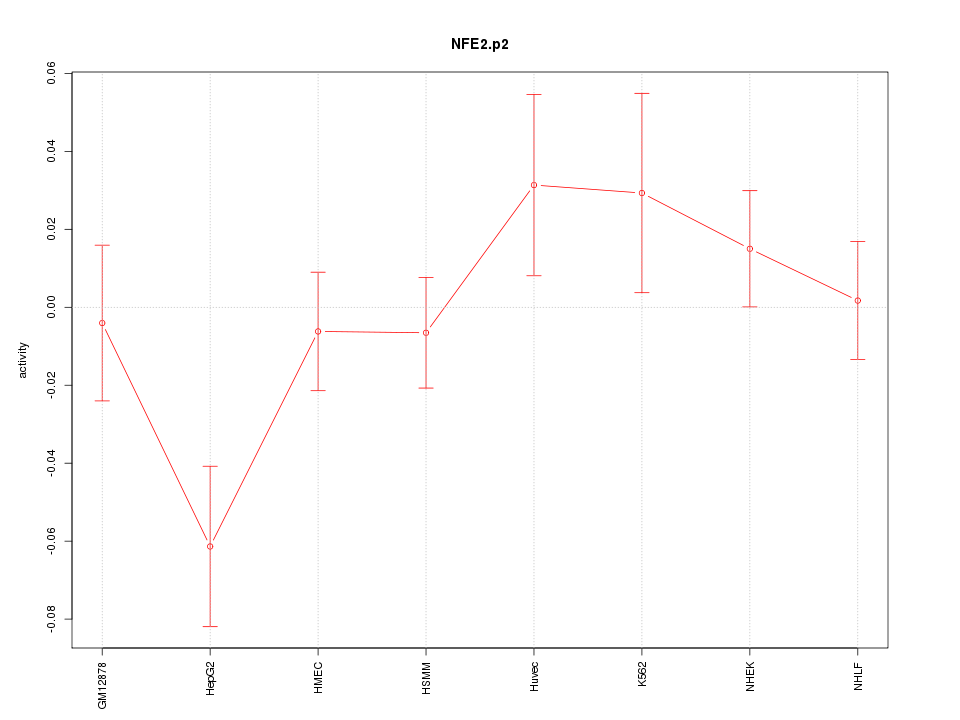
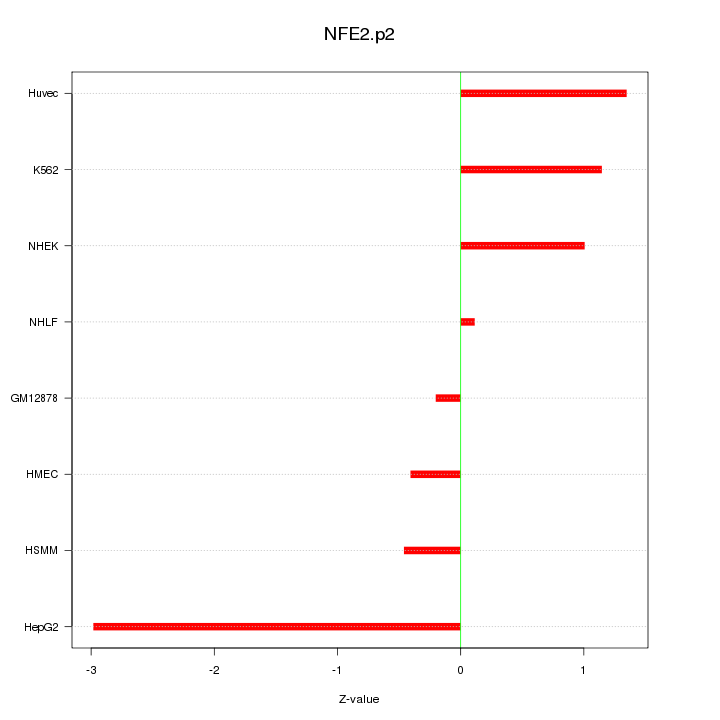
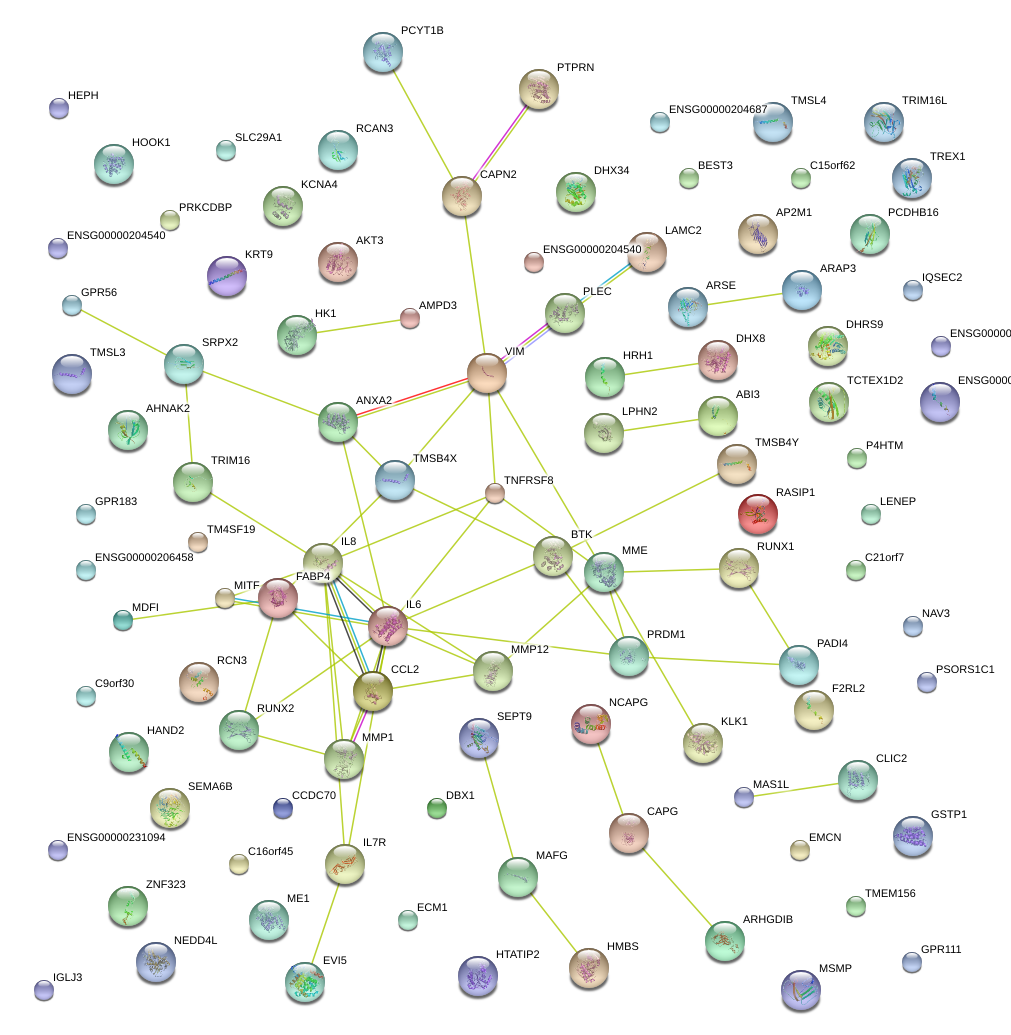
|
chr1_+_221955876
|
5.390
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr12_-_15005761
|
2.708
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr6_+_106653429
|
2.555
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr15_+_38849450
|
2.508
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr5_-_141041982
|
2.231
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr6_-_84197370
|
2.056
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr21_+_29439768
|
2.050
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr6_-_84197573
|
2.040
|
NM_002395
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr19_-_53935648
|
2.006
|
NM_017805
|
RASIP1
|
Ras interacting protein 1
|
|
chr6_-_84197463
|
1.989
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr11_+_20342262
|
1.967
|
NM_001098523
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr1_+_153232685
|
1.954
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr6_-_84197477
|
1.949
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr13_-_98757634
|
1.856
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr5_-_141041933
|
1.822
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr6_-_28429950
|
1.810
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr11_+_67107852
|
1.768
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr17_+_29606408
|
1.745
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr9_+_102231616
|
1.708
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr1_+_181421984
|
1.683
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_-_93030538
|
1.652
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr11_+_20341806
|
1.646
|
NM_006410
NM_001098521
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr11_+_20341835
|
1.635
|
NM_001098520
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr11_+_20341900
|
1.589
|
NM_001098522
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr11_-_6298284
|
1.529
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr15_-_58477407
|
1.516
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr17_+_18566060
|
1.515
|
NM_001037330
|
TRIM16L
|
tripartite motif containing 16-like
|
|
chr3_+_49002285
|
1.494
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr21_-_35343065
|
1.453
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_+_12903145
|
1.443
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr12_-_68369322
|
1.420
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chrX_-_154216929
|
1.401
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr11_+_67107641
|
1.399
|
NM_000852
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr1_-_242073094
|
1.385
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr16_+_56219802
|
1.383
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr11_+_10434056
|
1.381
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chrX_-_24575146
|
1.358
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr19_-_4510713
|
1.357
|
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chrX_-_24575375
|
1.335
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr1_+_148747110
|
1.332
|
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr3_+_69998064
|
1.330
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_-_38710376
|
1.316
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr1_+_148747208
|
1.290
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr13_+_51334117
|
1.288
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr18_+_54039779
|
1.284
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_+_22733290
|
1.283
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr2_+_169634330
|
1.269
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr1_+_12046020
|
1.266
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr22_+_21495273
|
1.265
|
|
|
|
|
chr18_+_54039730
|
1.246
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_54039818
|
1.245
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_+_11153778
|
1.232
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr17_+_72827185
|
1.219
|
NM_006640
|
SEPT9
|
septin 9
|
|
chr1_+_181422016
|
1.186
|
|
LAMC2
|
laminin, gamma 2
|
|
chr6_+_44302973
|
1.178
|
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr17_+_72827227
|
1.166
|
|
SEPT9
|
septin 9
|
|
chr1_+_181421796
|
1.163
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr2_-_219881937
|
1.159
|
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr6_+_41712907
|
1.158
|
|
MDFI
|
MyoD family inhibitor
|
|
chr5_-_75954807
|
1.152
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chrX_-_53327473
|
1.144
|
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chrX_-_100527799
|
1.115
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr17_-_77474597
|
1.114
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr17_+_44643067
|
1.101
|
|
ABI3
|
ABI family, member 3
|
|
chr11_+_118463906
|
1.084
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr6_+_31190505
|
1.074
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr6_-_29563657
|
1.067
|
NM_052967
|
MAS1L
|
MAS1 oncogene-like
|
|
chr8_-_145097031
|
1.066
|
NM_201380
|
PLEC
|
plectin
|
|
chrX_+_65299352
|
1.064
|
|
HEPH
|
hephaestin
|
|
chr1_+_81544432
|
1.063
|
|
LPHN2
|
latrophilin 2
|
|
chr3_+_156280129
|
1.056
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_+_17507276
|
1.051
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr3_-_197549640
|
1.034
|
NM_138461
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr11_-_102174032
|
1.032
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr14_-_104507862
|
1.010
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr11_-_102174102
|
1.002
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr4_+_74825138
|
0.996
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr3_+_48482213
|
0.995
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr8_-_82557956
|
0.995
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr3_+_185377341
|
0.995
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr3_+_48482625
|
0.984
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr4_-_101658094
|
0.980
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr3_-_197549588
|
0.980
|
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr2_-_85494664
|
0.974
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr12_+_76749199
|
0.970
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr16_+_15503623
|
0.961
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr2_-_85494639
|
0.961
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr9_-_35744271
|
0.955
|
NM_001044264
|
MSMP
|
microseminoprotein, prostate associated
|
|
chr22_+_21393107
|
0.954
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr4_-_174687104
|
0.954
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chrX_+_99786044
|
0.943
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chrX_-_2896285
|
0.930
|
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr5_+_35892747
|
0.929
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr10_+_17310263
|
0.926
|
NM_003380
|
VIM
|
vimentin
|
|
chr7_+_142205266
|
0.923
|
|
|
|
|
chr10_+_70745615
|
0.918
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr19_+_54723044
|
0.908
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr3_+_185377430
|
0.903
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr6_+_47732181
|
0.899
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chrX_+_69314060
|
0.898
|
NM_198512
|
DGAT2L6
|
diacylglycerol O-acyltransferase 2-like 6
|
|
chr20_+_33223434
|
0.888
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr1_-_115682374
|
0.885
|
NM_002506
|
NGF
|
nerve growth factor (beta polypeptide)
|
|
chr19_+_45564549
|
0.875
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr17_-_26665194
|
0.872
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr3_+_154035425
|
0.853
|
NM_002563
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1
|
|
chr5_+_150000394
|
0.850
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr1_-_156923111
|
0.841
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr15_+_99276982
|
0.835
|
NM_024652
|
LRRK1
|
leucine-rich repeat kinase 1
|
|
chr7_-_86526880
|
0.832
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr10_+_17310475
|
0.827
|
|
VIM
|
vimentin
|
|
chr17_-_26665213
|
0.805
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chrX_+_65299157
|
0.800
|
NM_138737
|
HEPH
|
hephaestin
|
|
chrX_-_15421279
|
0.799
|
NM_001018109
NM_003662
|
PIR
|
pirin (iron-binding nuclear protein)
|
|
chr22_-_25343959
|
0.798
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr9_+_35528890
|
0.795
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr8_-_42742729
|
0.787
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr1_+_226399026
|
0.786
|
NM_001159391
|
GUK1
|
guanylate kinase 1
|
|
chr21_-_35343331
|
0.774
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr22_-_25344037
|
0.770
|
|
CRYBB1
|
crystallin, beta B1
|
|
chr4_+_111053488
|
0.767
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr11_-_128399218
|
0.763
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr3_+_50259329
|
0.757
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr19_+_59107802
|
0.757
|
NM_031896
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7
|
|
chr8_-_122722674
|
0.755
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr22_-_43986896
|
0.755
|
|
KIAA0930
|
KIAA0930
|
|
chr17_+_27795598
|
0.754
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr1_+_24522116
|
0.753
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr4_-_5879488
|
0.748
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr9_+_35528628
|
0.743
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr4_+_154397975
|
0.743
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr17_+_27795645
|
0.738
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr19_-_55914837
|
0.734
|
|
|
|
|
chr17_-_26665144
|
0.733
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_+_27795633
|
0.730
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr17_+_22823151
|
0.729
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr16_-_70763524
|
0.727
|
NM_031293
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr6_+_34314545
|
0.723
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_+_27795635
|
0.722
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr3_+_66094324
|
0.713
|
NM_173471
|
SLC25A26
|
solute carrier family 25, member 26
|
|
chrX_+_100361588
|
0.713
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr11_-_66092534
|
0.707
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr3_-_193609531
|
0.701
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr1_+_12698704
|
0.700
|
NM_001103169
NM_001103170
|
AADACL3
|
arylacetamide deacetylase-like 3
|
|
chr7_-_128330615
|
0.699
|
|
KCP
|
kielin/chordin-like protein
|
|
chr22_+_20715340
|
0.699
|
|
|
|
|
chr19_+_43572679
|
0.695
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr12_-_90097384
|
0.694
|
NM_133503
|
DCN
|
decorin
|
|
chr3_+_185520744
|
0.694
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_8110268
|
0.693
|
|
C3AR1
|
complement component 3a receptor 1
|
|
chrX_+_99785818
|
0.691
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr21_-_35343428
|
0.683
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_+_185499679
|
0.682
|
NM_002808
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2
|
|
chr14_-_90780604
|
0.676
|
NM_001177676
|
GPR68
|
G protein-coupled receptor 68
|
|
chr7_+_150065594
|
0.673
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr12_-_101115677
|
0.671
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr15_+_76345577
|
0.669
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr3_+_101720753
|
0.669
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr11_-_47502115
|
0.661
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr4_-_68678181
|
0.655
|
NM_207407
|
TMPRSS11F
|
transmembrane protease, serine 11F
|
|
chr12_-_53099269
|
0.653
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr7_-_38355949
|
0.651
|
|
|
|
|
chr17_-_37022465
|
0.651
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr9_-_116920232
|
0.650
|
NM_002160
|
TNC
|
tenascin C
|
|
chr11_-_71458739
|
0.649
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr17_-_26665220
|
0.644
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr22_+_21464980
|
0.644
|
|
|
|
|
chr19_+_3317575
|
0.637
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chrX_-_33139349
|
0.636
|
NM_004006
|
DMD
|
dystrophin
|
|
chr15_+_38318528
|
0.636
|
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr17_+_27795632
|
0.625
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr21_-_35343500
|
0.622
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr15_-_74091817
|
0.620
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr12_-_119172339
|
0.609
|
NM_025157
|
PXN
|
paxillin
|
|
chr5_-_169657312
|
0.609
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_+_27795607
|
0.605
|
NM_002815
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr21_-_35343456
|
0.604
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr18_-_51219880
|
0.602
|
|
TCF4
|
transcription factor 4
|
|
chr17_-_44642934
|
0.601
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr3_-_15814678
|
0.597
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_-_109770474
|
0.596
|
NM_002790
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr22_+_21340757
|
0.594
|
|
|
|
|
chrX_+_133952633
|
0.593
|
NM_001163438
|
LOC644538
|
hypothetical protein LOC644538
|
|
chr1_-_119333701
|
0.590
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr5_-_169657192
|
0.589
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_-_74244539
|
0.588
|
|
|
|
|
chr1_-_109770581
|
0.588
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr5_-_169657395
|
0.587
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_-_35975227
|
0.586
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr2_-_65447370
|
0.584
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr1_-_109770567
|
0.584
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr19_+_3317536
|
0.584
|
NM_005597
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chrX_-_72351348
|
0.583
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr3_+_70131572
|
0.583
|
|
|
|
|
chr10_+_85889164
|
0.583
|
NM_014394
|
GHITM
|
growth hormone inducible transmembrane protein
|
|
chr17_-_39700960
|
0.582
|
NM_000342
|
SLC4A1
|
solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group)
|
|
chr12_-_53099277
|
0.579
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr3_+_194441586
|
0.578
|
NM_020386
|
HRASLS
|
HRAS-like suppressor
|
|
chr9_+_130231027
|
0.578
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr22_-_43987010
|
0.573
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr15_+_38319326
|
0.571
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|