|
chr2_+_219453487
|
8.592
|
NM_025216
|
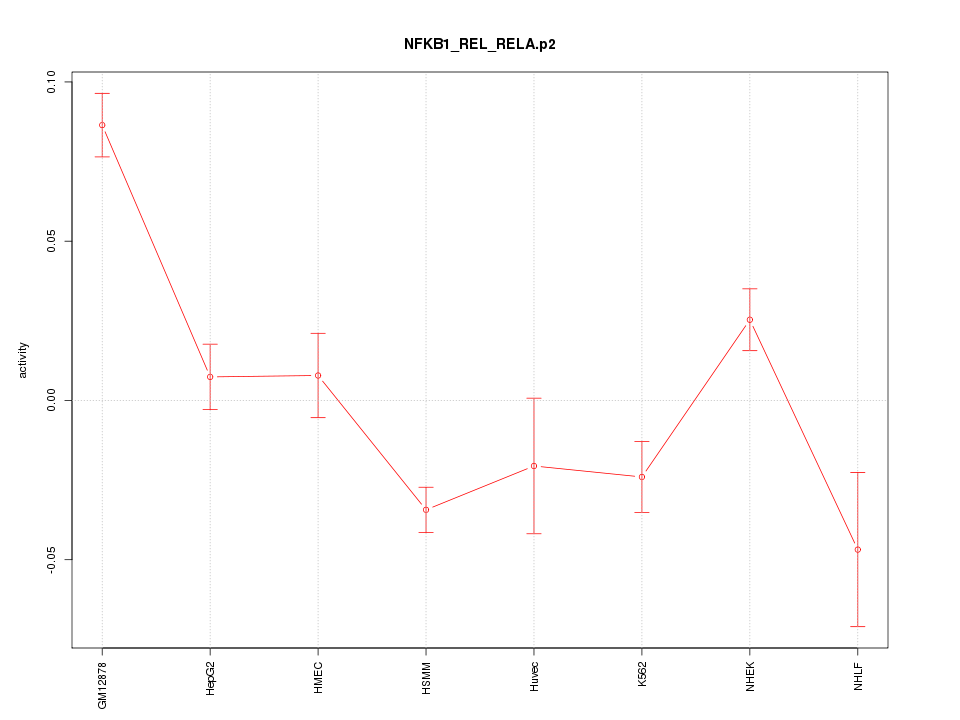
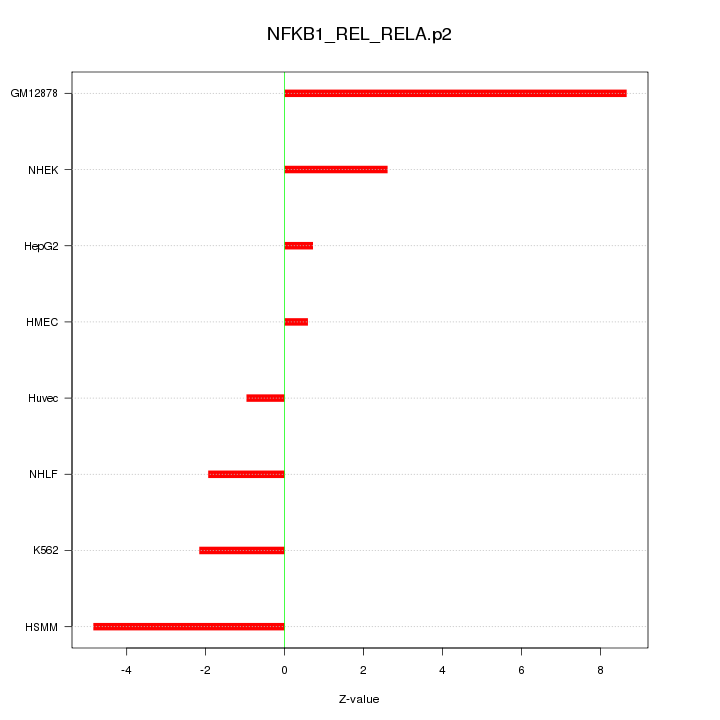
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr2_-_231498184
|
5.631
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr17_+_35277942
|
5.306
|
NM_198844
NM_199321
|
ZPBP2
|
zona pellucida binding protein 2
|
|
chr20_-_4743768
|
5.296
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr17_-_33178987
|
5.047
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr19_+_4180494
|
4.969
|
NM_005755
|
EBI3
|
Epstein-Barr virus induced 3
|
|
chr9_+_126094067
|
4.602
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr14_-_95250155
|
4.435
|
NM_001098725
NM_021966
|
TCL1A
|
T-cell leukemia/lymphoma 1A
|
|
chr3_-_13089547
|
4.127
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr12_+_6925997
|
4.087
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr19_-_6542012
|
3.952
|
|
CD70
|
CD70 molecule
|
|
chr19_+_40632456
|
3.935
|
NM_005306
|
FFAR2
|
free fatty acid receptor 2
|
|
chr9_+_126094388
|
3.863
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr12_+_6926014
|
3.819
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr20_-_44370537
|
3.738
|
|
CDH22
|
cadherin 22, type 2
|
|
chr19_-_7672978
|
3.735
|
NM_002002
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr17_+_45993338
|
3.651
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr17_+_7402315
|
3.572
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr15_+_79376295
|
3.378
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr17_+_22823151
|
3.366
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr15_-_38185578
|
3.344
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr19_+_47073029
|
3.340
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr1_-_24386324
|
3.308
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr6_+_32713160
|
3.271
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr3_+_123279439
|
3.208
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr1_+_6767968
|
3.197
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr22_-_35875890
|
3.087
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr3_-_58175437
|
3.005
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr19_+_3087026
|
2.992
|
NM_002068
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class)
|
|
chr1_+_26744929
|
2.916
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr11_-_60880799
|
2.893
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr1_-_200397300
|
2.871
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr6_-_30188845
|
2.827
|
NM_007028
|
TRIM31
|
tripartite motif containing 31
|
|
chr3_-_113334657
|
2.827
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr17_-_16816113
|
2.823
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr2_-_74522563
|
2.795
|
NM_001015055
|
RTKN
|
rhotekin
|
|
chr1_+_26744893
|
2.786
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr17_-_33179118
|
2.722
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr14_-_69725342
|
2.703
|
NM_033262
NM_058240
NM_182932
NM_183002
|
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3
|
|
chr2_-_74522534
|
2.689
|
|
RTKN
|
rhotekin
|
|
chr6_+_31648071
|
2.680
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr2_-_74522513
|
2.677
|
|
RTKN
|
rhotekin
|
|
chr6_+_31647854
|
2.647
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr11_-_65070625
|
2.644
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr19_+_17499064
|
2.620
|
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr17_-_35506493
|
2.611
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr17_-_31231421
|
2.584
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr6_+_30958672
|
2.573
|
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr14_+_75058518
|
2.570
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr13_-_49597776
|
2.560
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr1_-_200395741
|
2.542
|
NM_080588
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr16_+_49288550
|
2.527
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr5_-_149772528
|
2.517
|
|
|
|
|
chr7_-_44985084
|
2.471
|
|
MYO1G
|
myosin IG
|
|
chrX_-_30505804
|
2.464
|
NM_025159
|
CXorf21
|
chromosome X open reading frame 21
|
|
chr14_+_75058591
|
2.415
|
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr12_+_50731457
|
2.388
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr1_+_67404756
|
2.386
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr18_-_6404900
|
2.385
|
NM_173464
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila)
|
|
chrX_+_12795111
|
2.350
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr14_-_23878871
|
2.321
|
NM_006871
|
RIPK3
|
receptor-interacting serine-threonine kinase 3
|
|
chr17_-_58877276
|
2.321
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr1_-_159275334
|
2.311
|
|
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
|
|
chr6_-_32659958
|
2.308
|
|
HLA-DRB1
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 5
|
|
chr1_-_207892420
|
2.307
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr2_+_233633348
|
2.296
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr12_+_111980044
|
2.268
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr19_+_5082230
|
2.265
|
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr1_+_25816545
|
2.242
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr19_-_48976852
|
2.226
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr2_+_68815416
|
2.223
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr5_-_149772469
|
2.200
|
NM_001025159
NM_004355
NM_001025158
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr2_+_233633279
|
2.193
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr12_-_53661888
|
2.176
|
NM_001098815
|
KIAA0748
|
KIAA0748
|
|
chr19_+_1018536
|
2.173
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr11_+_66580884
|
2.158
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr15_+_57517564
|
2.146
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr5_+_60663842
|
2.142
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr1_-_181826633
|
2.127
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr19_-_48977247
|
2.122
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr19_-_6542082
|
2.119
|
|
CD70
|
CD70 molecule
|
|
chr17_+_37694007
|
2.110
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr2_+_237143118
|
2.105
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr6_-_27387989
|
2.101
|
NM_033482
|
POM121L2
|
POM121 membrane glycoprotein-like 2
|
|
chr17_+_8865547
|
2.082
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr10_-_6144119
|
2.060
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr20_+_44180296
|
2.051
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr17_-_71352182
|
2.043
|
NM_199242
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr19_+_17723285
|
2.037
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr20_-_55628852
|
2.018
|
NM_001160417
NM_001160418
NM_001160419
NM_030776
|
ZBP1
|
Z-DNA binding protein 1
|
|
chr19_+_44697592
|
1.976
|
NM_182704
|
SELV
|
selenoprotein V
|
|
chr10_-_134940315
|
1.971
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr1_+_200246370
|
1.967
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr6_-_29635456
|
1.966
|
|
UBD
|
ubiquitin D
|
|
chr19_-_47190160
|
1.946
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr12_+_47658502
|
1.942
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr22_-_35970133
|
1.925
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr11_+_120828058
|
1.919
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr16_+_84490267
|
1.917
|
NM_002163
|
IRF8
|
interferon regulatory factor 8
|
|
chr11_+_66580861
|
1.915
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr2_+_231968578
|
1.913
|
NM_145236
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7
|
|
chr19_+_55614006
|
1.896
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr1_+_154389945
|
1.896
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr22_-_35970230
|
1.868
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr4_-_120768563
|
1.866
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr18_-_26996742
|
1.852
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr9_-_139235595
|
1.849
|
NM_031297
|
RNF208
|
ring finger protein 208
|
|
chr1_-_159275357
|
1.832
|
NM_001113205
NM_001113206
NM_001113207
|
TSTD1
F11R
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
F11 receptor
|
|
chr10_+_102746819
|
1.829
|
NM_032429
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr5_+_177473071
|
1.824
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr15_-_72804763
|
1.818
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr7_+_149042960
|
1.817
|
NM_032534
|
KRBA1
|
KRAB-A domain containing 1
|
|
chr1_-_200396354
|
1.816
|
NM_002832
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr1_+_154390939
|
1.814
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr1_-_200396328
|
1.794
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr19_-_17819814
|
1.789
|
NM_000215
|
JAK3
|
Janus kinase 3
|
|
chr7_-_44985202
|
1.788
|
NM_033054
|
MYO1G
|
myosin IG
|
|
chr1_-_233879617
|
1.787
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr6_-_29635680
|
1.783
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr17_-_71352017
|
1.768
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr6_+_14225932
|
1.765
|
|
CD83
|
CD83 molecule
|
|
chr5_-_149772563
|
1.754
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr19_-_17819773
|
1.754
|
|
JAK3
|
Janus kinase 3
|
|
chr19_-_4289750
|
1.739
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr8_-_70909754
|
1.739
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr13_-_45654327
|
1.724
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_199130571
|
1.709
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr1_-_205210567
|
1.708
|
NM_001122979
NM_001170631
NM_032029
|
FCAMR
|
Fc receptor, IgA, IgM, high affinity
|
|
chr16_+_66128843
|
1.705
|
NM_001193524
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr1_+_200246312
|
1.701
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr5_+_176807931
|
1.692
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr11_+_118259684
|
1.690
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr1_-_181826292
|
1.688
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr8_-_22606401
|
1.687
|
|
EGR3
|
early growth response 3
|
|
chr9_-_139068269
|
1.677
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr17_+_69974116
|
1.674
|
NM_007261
|
CD300A
|
CD300a molecule
|
|
chrX_+_78087484
|
1.668
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr19_+_8180179
|
1.623
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr8_-_11096257
|
1.605
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr8_+_144706699
|
1.602
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr7_-_44985175
|
1.588
|
|
MYO1G
|
myosin IG
|
|
chr12_-_28016182
|
1.571
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr9_+_139047441
|
1.571
|
|
C9orf139
|
chromosome 9 open reading frame 139
|
|
chr7_-_150737980
|
1.555
|
NM_198285
|
WDR86
|
WD repeat domain 86
|
|
chr14_-_50631407
|
1.552
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr5_+_96319910
|
1.552
|
NM_175920
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr18_-_26935427
|
1.542
|
|
DSC2
|
desmocollin 2
|
|
chr3_+_113200696
|
1.541
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr11_+_122031549
|
1.523
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr4_+_107036076
|
1.523
|
|
NPNT
|
nephronectin
|
|
chr8_-_11361594
|
1.504
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr16_-_55875953
|
1.502
|
|
PLLP
|
plasmolipin
|
|
chr4_-_13158391
|
1.491
|
|
LOC285548
|
hypothetical LOC285548
|
|
chr16_-_73798539
|
1.489
|
NM_001025200
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr17_+_45401560
|
1.459
|
NM_138281
|
DLX4
|
distal-less homeobox 4
|
|
chr11_-_133332089
|
1.454
|
NM_014987
|
IGSF9B
|
immunoglobulin superfamily, member 9B
|
|
chr17_+_7402788
|
1.451
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr1_-_164402700
|
1.446
|
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr4_-_77163609
|
1.440
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr1_-_21489235
|
1.409
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr19_-_11549441
|
1.404
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr14_-_23106779
|
1.394
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr22_+_38720881
|
1.393
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr16_-_55876075
|
1.391
|
NM_015993
|
PLLP
|
plasmolipin
|
|
chr9_-_135334003
|
1.383
|
NM_001145099
NM_017585
|
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chr16_-_31135895
|
1.382
|
NM_152901
|
PYDC1
|
PYD (pyrin domain) containing 1
|
|
chr16_+_23221091
|
1.379
|
NM_000336
|
SCNN1B
|
sodium channel, nonvoltage-gated 1, beta
|
|
chr5_+_10617431
|
1.377
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr7_+_68702516
|
1.377
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr9_+_126579203
|
1.367
|
NM_182487
|
OLFML2A
|
olfactomedin-like 2A
|
|
chr16_-_55876099
|
1.352
|
|
PLLP
|
plasmolipin
|
|
chr1_-_153107084
|
1.349
|
|
|
|
|
chr19_+_10388448
|
1.345
|
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr17_+_21131940
|
1.328
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr11_+_120828275
|
1.325
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr12_-_6611075
|
1.325
|
NM_001142961
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr16_+_28903647
|
1.321
|
NM_001014989
|
LAT
|
linker for activation of T cells
|
|
chr20_-_24986501
|
1.314
|
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr15_+_74139325
|
1.307
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr1_-_7923464
|
1.306
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr1_+_35104120
|
1.302
|
|
|
|
|
chr2_+_95055155
|
1.295
|
NM_002371
NM_022438
NM_022439
NM_022440
|
MAL
|
mal, T-cell differentiation protein
|
|
chr10_-_98263434
|
1.295
|
NM_012465
|
TLL2
|
tolloid-like 2
|
|
chr5_-_157011949
|
1.287
|
NM_007017
NM_178424
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr19_+_41039649
|
1.283
|
NM_032123
NM_199179
NM_199180
|
KIRREL2
|
kin of IRRE like 2 (Drosophila)
|
|
chr8_-_95343716
|
1.276
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr19_-_56214765
|
1.275
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr2_-_108972100
|
1.273
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr19_-_6542136
|
1.271
|
NM_001252
|
CD70
|
CD70 molecule
|
|
chr19_+_13066956
|
1.267
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr17_-_1336073
|
1.261
|
|
MYO1C
|
myosin IC
|
|
chr20_+_44180355
|
1.260
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr1_-_92721633
|
1.253
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr9_-_116608113
|
1.251
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr10_-_61139328
|
1.239
|
NM_194298
|
LOC100129721
SLC16A9
|
hypothetical protein LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr1_-_53159759
|
1.238
|
|
ECHDC2
|
enoyl CoA hydratase domain containing 2
|
|
chr19_-_4491024
|
1.238
|
NM_052972
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr1_-_27834210
|
1.231
|
NM_005248
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr1_-_92721943
|
1.220
|
NM_001127215
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr3_+_195889845
|
1.212
|
|
FAM43A
|
family with sequence similarity 43, member A
|