|
chr11_-_85457469
|
1.825
|
|
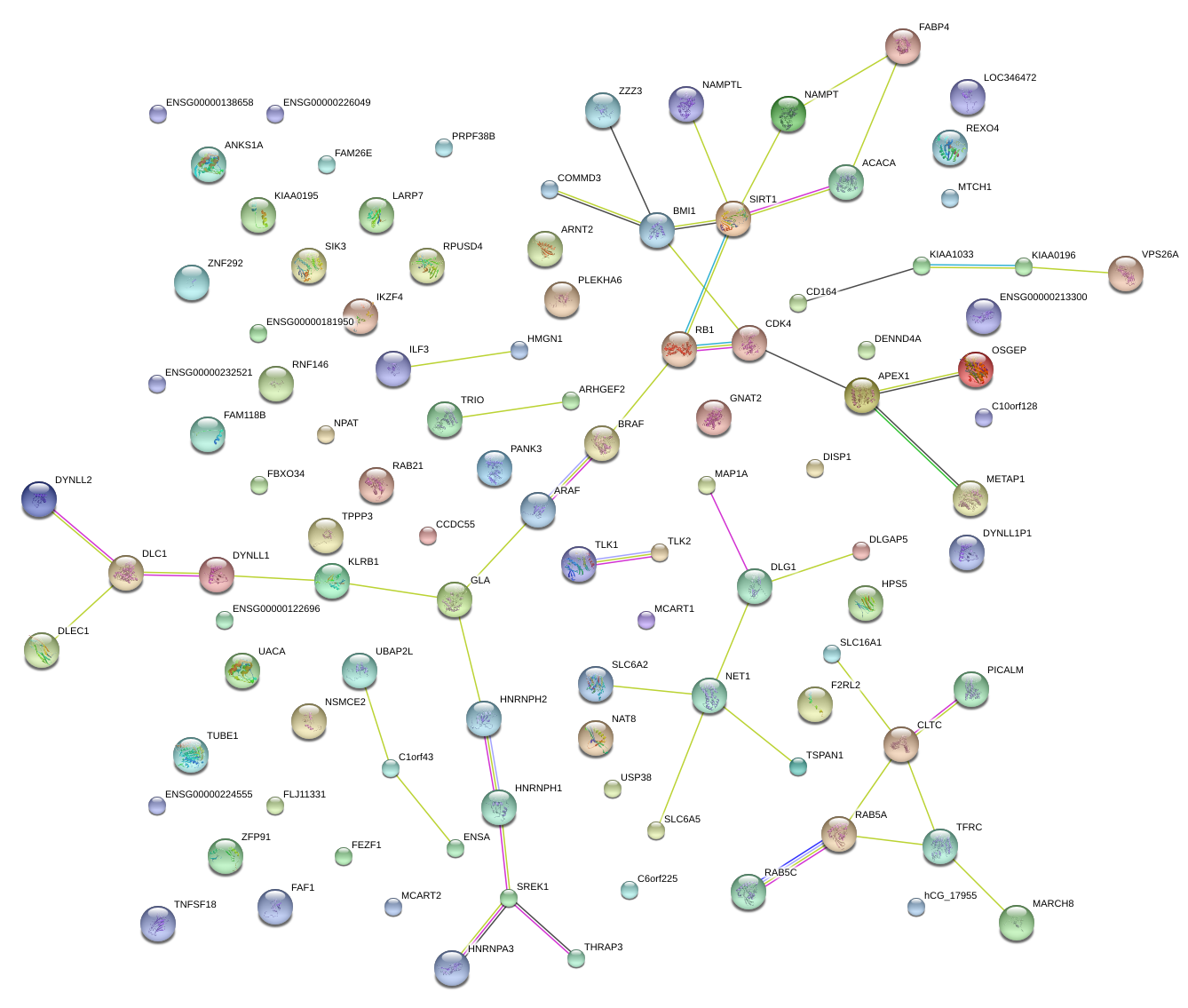
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr15_-_68781662
|
1.389
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr11_-_85457736
|
1.004
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr11_-_85457753
|
0.948
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_-_105712565
|
0.846
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr4_-_113777569
|
0.825
|
NM_018392
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr8_+_126173264
|
0.794
|
NM_173685
|
NSMCE2
|
non-SMC element 2, MMS21 homolog (S. cerevisiae)
|
|
chr15_+_41597097
|
0.786
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr17_+_55051818
|
0.780
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr3_+_19963534
|
0.766
|
NM_004162
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr12_-_56432377
|
0.757
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_+_221168388
|
0.742
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr11_+_58103212
|
0.740
|
|
ZFP91-CNTF
ZFP91
|
ZFP91-CNTF readthrough transcript
zinc finger protein 91 homolog (mouse)
|
|
chr8_-_126173108
|
0.736
|
|
KIAA0196
|
KIAA0196
|
|
chr12_-_56432375
|
0.729
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432385
|
0.728
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr13_+_47776026
|
0.724
|
|
RB1
|
retinoblastoma 1
|
|
chr1_-_77920914
|
0.719
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr1_-_154205675
|
0.713
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr8_-_126173186
|
0.699
|
|
KIAA0196
|
KIAA0196
|
|
chr6_-_37061920
|
0.683
|
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr19_+_10626106
|
0.679
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr13_+_47775951
|
0.678
|
|
RB1
|
retinoblastoma 1
|
|
chr14_+_54807855
|
0.674
|
|
FBXO34
|
F-box protein 34
|
|
chr6_-_109810396
|
0.663
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr12_+_104025667
|
0.662
|
|
KIAA1033
|
KIAA1033
|
|
chr17_+_70984173
|
0.661
|
|
KIAA0195
|
KIAA0195
|
|
chr8_-_126173203
|
0.657
|
|
KIAA0196
|
KIAA0196
|
|
chr8_-_13416554
|
0.656
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_-_126173189
|
0.655
|
|
KIAA0196
|
KIAA0196
|
|
chr12_+_104025675
|
0.650
|
|
KIAA1033
|
KIAA1033
|
|
chrX_+_100549885
|
0.648
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr5_-_167938875
|
0.646
|
|
PANK3
|
pantothenate kinase 3
|
|
chr10_+_5478521
|
0.641
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr1_+_36462625
|
0.638
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr15_-_63871492
|
0.638
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr3_-_198509276
|
0.627
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr5_-_75954807
|
0.619
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr12_-_56432292
|
0.615
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr4_+_144325502
|
0.615
|
NM_032557
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr4_-_113777470
|
0.600
|
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr7_-_105712646
|
0.600
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr17_-_32730804
|
0.596
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr6_+_87921980
|
0.593
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr14_-_19993032
|
0.585
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr7_-_121731721
|
0.582
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr1_-_113300138
|
0.582
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr17_+_55052046
|
0.581
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chrX_-_100549561
|
0.574
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr1_+_36462583
|
0.572
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr6_+_127629677
|
0.572
|
NM_030963
|
RNF146
|
ring finger protein 146
|
|
chr4_+_113777568
|
0.567
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr1_-_51198328
|
0.563
|
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr4_+_144325533
|
0.563
|
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr8_-_126173055
|
0.562
|
|
KIAA0196
|
KIAA0196
|
|
chr5_+_14493905
|
0.558
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr17_+_55052060
|
0.556
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr1_-_51198383
|
0.553
|
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr12_+_54700955
|
0.552
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chrX_-_100549444
|
0.551
|
|
GLA
|
galactosidase, alpha
|
|
chr6_-_37061908
|
0.550
|
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr1_-_171286634
|
0.549
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr13_+_47775901
|
0.546
|
|
RB1
|
retinoblastoma 1
|
|
chr6_-_37061897
|
0.545
|
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr1_-_109957195
|
0.544
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr8_-_126173227
|
0.541
|
NM_014846
|
KIAA0196
|
KIAA0196
|
|
chr1_-_51198417
|
0.538
|
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr17_+_25467948
|
0.524
|
NM_032141
|
CCDC55
|
coiled-coil domain containing 55
|
|
chr6_+_34964999
|
0.521
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr1_-_51198506
|
0.520
|
NM_007051
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr6_-_37061902
|
0.518
|
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr15_-_63871598
|
0.515
|
NM_001144823
NM_005848
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr12_+_104025578
|
0.513
|
NM_015275
|
KIAA1033
|
KIAA1033
|
|
chrX_+_47305459
|
0.511
|
NM_001654
|
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog
|
|
chr11_-_107598538
|
0.511
|
NM_002519
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chr10_+_69314714
|
0.511
|
|
SIRT1
|
sirtuin 1
|
|
chr12_+_70434927
|
0.510
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr1_-_202595666
|
0.508
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr16_-_65984836
|
0.507
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr11_+_125586828
|
0.492
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr9_-_37894098
|
0.491
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chrX_+_100549869
|
0.491
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr11_-_116474196
|
0.490
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr6_-_109810296
|
0.489
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr5_-_167938991
|
0.489
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr10_-_45410197
|
0.487
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr6_-_112515329
|
0.487
|
NM_016262
|
TUBE1
|
tubulin, epsilon 1
|
|
chr6_+_112515366
|
0.487
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr6_-_37061924
|
0.486
|
NM_014341
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chrX_+_100549839
|
0.484
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr5_+_65476418
|
0.483
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr12_+_104025686
|
0.482
|
|
|
|
|
chr6_+_116939500
|
0.481
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr7_-_140270749
|
0.478
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr10_+_70553936
|
0.478
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr2_+_177785667
|
0.476
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr1_+_152460381
|
0.474
|
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr11_-_18300246
|
0.473
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr10_-_50066386
|
0.471
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr21_-_39642712
|
0.469
|
|
HMGN1
|
high-mobility group nucleosome binding domain 1
|
|
chr12_-_44670475
|
0.469
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr8_-_82557956
|
0.467
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr6_-_109810310
|
0.461
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr10_+_22645370
|
0.460
|
|
COMMD3
|
COMM domain containing 3
|
|
chr14_+_19993181
|
0.458
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr1_-_152459506
|
0.457
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr11_-_107598472
|
0.456
|
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chr11_-_125586733
|
0.453
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr10_+_70553949
|
0.448
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chrX_+_100549861
|
0.447
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_-_148868535
|
0.446
|
|
ENSA
|
endosulfine alpha
|
|
chr10_+_70553945
|
0.445
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr3_-_197293331
|
0.445
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr14_+_54807773
|
0.444
|
NM_017943
|
FBXO34
|
F-box protein 34
|
|
chr2_-_171725604
|
0.440
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr5_+_15553768
|
0.440
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr10_+_70553993
|
0.440
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_+_76640607
|
0.436
|
|
VDAC2
|
voltage-dependent anion channel 2
|
|
chr3_-_197293377
|
0.431
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr12_-_113330574
|
0.429
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr12_-_8993460
|
0.428
|
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr1_-_115060904
|
0.427
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr3_-_99724373
|
0.427
|
|
CLDND1
|
claudin domain containing 1
|
|
chr15_-_32289561
|
0.426
|
NM_024713
|
C15orf29
|
chromosome 15 open reading frame 29
|
|
chr6_-_109810354
|
0.424
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr3_-_99724047
|
0.423
|
|
CLDND1
|
claudin domain containing 1
|
|
chr6_-_79844658
|
0.423
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr10_+_70553962
|
0.420
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr6_-_49712483
|
0.416
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr11_+_18300370
|
0.413
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr10_-_74055824
|
0.410
|
|
CBARA1
|
calcium binding atopy-related autoantigen 1
|
|
chr3_+_19963865
|
0.409
|
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr1_-_51198052
|
0.408
|
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr4_-_977163
|
0.408
|
NM_022042
NM_134425
NM_213613
|
SLC26A1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr5_-_16795336
|
0.407
|
|
MYO10
|
myosin X
|
|
chr3_-_197293355
|
0.407
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr12_+_50592379
|
0.405
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr3_-_197293394
|
0.404
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr3_+_134239860
|
0.402
|
NM_001136469
NM_023943
|
TMEM108
|
transmembrane protein 108
|
|
chr2_+_105320440
|
0.401
|
NM_024093
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr12_-_90097384
|
0.396
|
NM_133503
|
DCN
|
decorin
|
|
chr11_+_107598768
|
0.396
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr3_+_159845067
|
0.395
|
|
GFM1
|
G elongation factor, mitochondrial 1
|
|
chr1_-_148868644
|
0.392
|
|
ENSA
|
endosulfine alpha
|
|
chr5_-_90645974
|
0.392
|
|
LOC100505994
|
hypothetical LOC100505994
|
|
chr3_+_134239906
|
0.391
|
|
TMEM108
|
transmembrane protein 108
|
|
chr12_-_90096275
|
0.390
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chrX_+_101862297
|
0.389
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr2_-_177836773
|
0.389
|
NM_001145412
NM_001145413
|
NFE2L2
|
nuclear factor (erythroid-derived 2)-like 2
|
|
chr1_+_52294554
|
0.388
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr9_+_102380156
|
0.388
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr3_+_185515108
|
0.386
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr17_-_54538816
|
0.386
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr1_-_70443748
|
0.384
|
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr6_+_143423656
|
0.384
|
NM_016108
|
AIG1
|
androgen-induced 1
|
|
chr1_-_203985701
|
0.383
|
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr3_-_12680329
|
0.383
|
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr22_+_39583001
|
0.383
|
NM_022098
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative
|
|
chr17_-_54538896
|
0.382
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr9_-_124707314
|
0.381
|
NM_001100588
NM_018835
|
RC3H2
|
ring finger and CCCH-type domains 2
|
|
chr1_-_203985752
|
0.380
|
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr1_-_227828416
|
0.380
|
NM_001025247
NM_014409
|
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr1_-_152797760
|
0.380
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr6_-_109810246
|
0.379
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr1_-_152797707
|
0.379
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr3_+_52694975
|
0.379
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr12_+_34066679
|
0.378
|
|
ALG10
|
asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe)
|
|
chr1_-_203985780
|
0.378
|
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr1_+_220952518
|
0.377
|
NM_144695
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr12_+_99185289
|
0.375
|
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr12_+_99185334
|
0.375
|
NM_017988
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr1_-_148868664
|
0.374
|
|
ENSA
|
endosulfine alpha
|
|
chr12_-_69317441
|
0.372
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr6_+_144040751
|
0.370
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr9_-_90983195
|
0.369
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr1_+_181871830
|
0.369
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr17_+_55052065
|
0.369
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr12_-_8993515
|
0.369
|
NM_002355
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr1_-_148868713
|
0.369
|
NM_004436
NM_207042
NM_207043
NM_207044
NM_207168
|
ENSA
|
endosulfine alpha
|
|
chr1_-_33055431
|
0.368
|
|
YARS
|
tyrosyl-tRNA synthetase
|
|
chr10_+_70553983
|
0.368
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_+_22645304
|
0.367
|
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 read-through transcript
COMM domain containing 3
|
|
chr4_+_175441402
|
0.367
|
NM_001040157
|
KIAA1712
|
KIAA1712
|
|
chr12_+_70434920
|
0.367
|
NM_014999
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr14_-_21014871
|
0.366
|
NM_032846
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr10_-_74055859
|
0.366
|
|
CBARA1
|
calcium binding atopy-related autoantigen 1
|
|
chr1_-_148868671
|
0.366
|
|
ENSA
|
endosulfine alpha
|
|
chr5_-_94916416
|
0.365
|
NM_014639
|
TTC37
|
tetratricopeptide repeat domain 37
|
|
chr1_+_52294563
|
0.364
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr14_+_19993129
|
0.363
|
NM_001641
NM_080648
NM_080649
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr12_-_122415327
|
0.363
|
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr1_+_220952681
|
0.362
|
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr10_+_76640522
|
0.361
|
|
VDAC2
|
voltage-dependent anion channel 2
|
|
chrX_-_48699702
|
0.361
|
|
OTUD5
|
OTU domain containing 5
|
|
chr16_+_23754822
|
0.360
|
|
PRKCB
|
protein kinase C, beta
|
|
chr6_-_32121784
|
0.360
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr1_+_1994943
|
0.359
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr19_-_9908014
|
0.358
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr1_+_10015736
|
0.357
|
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr6_+_109437599
|
0.356
|
|
|
|