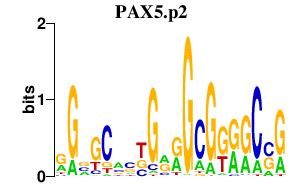
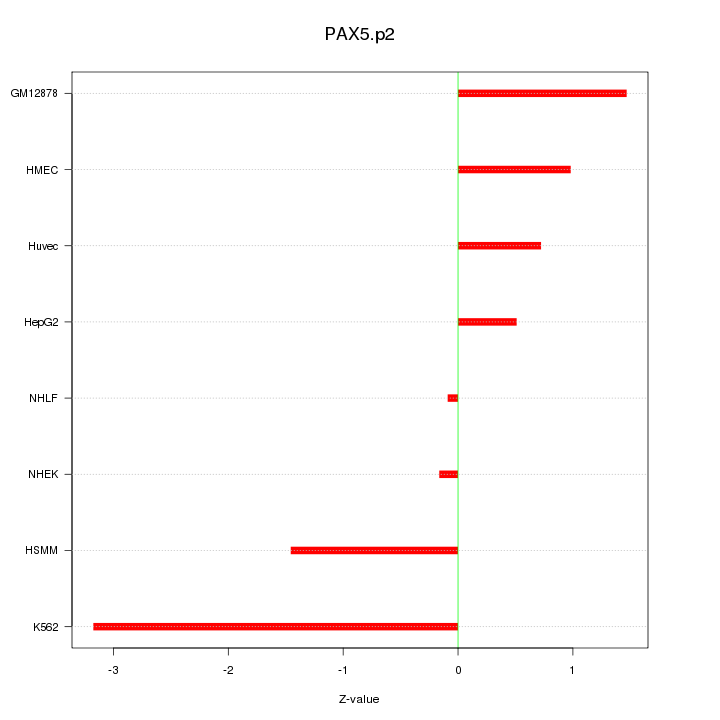
|
chr1_-_233879617
|
4.317
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr16_-_66827449
|
3.046
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr13_-_43351825
|
2.854
|
NM_144974
|
CCDC122
|
coiled-coil domain containing 122
|
|
chr19_+_8180179
|
2.424
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr13_+_43351968
|
2.406
|
NM_153218
|
C13orf31
|
chromosome 13 open reading frame 31
|
|
chr2_-_36384
|
2.379
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr19_+_61742111
|
2.334
|
NM_020828
|
ZFP28
|
zinc finger protein 28 homolog (mouse)
|
|
chr7_-_107883929
|
2.036
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr10_+_135042684
|
2.028
|
NM_152911
NM_207127
NM_207128
|
PAOX
|
polyamine oxidase (exo-N4-amino)
|
|
chr2_-_127694123
|
1.984
|
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr15_-_50375143
|
1.895
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr14_-_94855848
|
1.876
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr12_-_93568336
|
1.793
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr5_+_17270474
|
1.746
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr16_-_265868
|
1.730
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr15_+_99237527
|
1.725
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr10_-_30064600
|
1.665
|
NM_003174
|
SVIL
|
supervillin
|
|
chr17_+_27617307
|
1.594
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr2_-_88533167
|
1.591
|
NM_001135649
|
FOXI3
|
forkhead box I3
|
|
chr9_-_91302707
|
1.552
|
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr11_-_60819273
|
1.532
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr10_-_105604942
|
1.522
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr2_-_98714020
|
1.509
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr5_+_175725050
|
1.502
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr20_-_38751289
|
1.499
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr10_-_71575953
|
1.489
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr13_+_43845977
|
1.464
|
NM_001010897
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2
|
|
chr10_+_131155443
|
1.458
|
NM_002412
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr1_+_61980679
|
1.435
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr10_+_131155490
|
1.433
|
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr5_+_17270781
|
1.432
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr17_-_59131194
|
1.430
|
|
LIMD2
|
LIM domain containing 2
|
|
chr3_+_32255158
|
1.425
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr1_-_2333800
|
1.390
|
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr4_-_7991992
|
1.379
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr19_-_47619992
|
1.343
|
|
LIPE
|
lipase, hormone-sensitive
|
|
chr18_-_68685789
|
1.336
|
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr14_-_99842519
|
1.324
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr11_+_8059267
|
1.314
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr6_-_89884324
|
1.305
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr22_-_49317408
|
1.293
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr4_-_809879
|
1.283
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr1_+_38032045
|
1.278
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr20_+_1154763
|
1.275
|
NM_001136566
|
RAD21L1
|
RAD21-like 1 (S. pombe)
|
|
chr14_-_91372365
|
1.264
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr19_-_19599973
|
1.263
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr11_-_60819110
|
1.261
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr19_-_242335
|
1.257
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr10_+_42892506
|
1.211
|
NM_020630
NM_020975
|
RET
|
ret proto-oncogene
|
|
chr5_+_43228083
|
1.209
|
NM_153361
|
MGC42105
|
serine/threonine-protein kinase NIM1
|
|
chr1_-_2333863
|
1.200
|
NM_002617
NM_153818
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr9_+_138497767
|
1.195
|
NM_152571
|
C9orf163
|
chromosome 9 open reading frame 163
|
|
chr5_+_68824143
|
1.185
|
|
OCLN
|
occludin
|
|
chr13_+_34414390
|
1.179
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr17_+_40565492
|
1.171
|
NM_001135704
|
ACBD4
|
acyl-CoA binding domain containing 4
|
|
chr7_-_117300763
|
1.165
|
NM_033427
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr3_+_32255215
|
1.161
|
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr14_+_102128741
|
1.154
|
|
RCOR1
|
REST corepressor 1
|
|
chr5_+_139155569
|
1.145
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr5_+_96023926
|
1.144
|
|
CAST
|
calpastatin
|
|
chr14_+_93710401
|
1.143
|
NM_020958
NM_058237
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4
|
|
chr22_-_36153449
|
1.143
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr21_+_32706615
|
1.140
|
NM_058187
|
C21orf63
|
chromosome 21 open reading frame 63
|
|
chr8_-_26427304
|
1.133
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr8_+_26427378
|
1.124
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr13_+_43351419
|
1.118
|
NM_001128303
|
C13orf31
|
chromosome 13 open reading frame 31
|
|
chr3_-_49289285
|
1.116
|
NM_198562
|
C3orf62
|
chromosome 3 open reading frame 62
|
|
chr2_-_98713862
|
1.107
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr3_+_133923758
|
1.102
|
|
NCRNA00119
|
non-protein coding RNA 119
|
|
chr3_-_53054999
|
1.086
|
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr16_+_65790514
|
1.077
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr2_+_231968578
|
1.074
|
NM_145236
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7
|
|
chrX_+_43399101
|
1.073
|
|
MAOA
|
monoamine oxidase A
|
|
chr19_+_39664706
|
1.070
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr8_+_25097983
|
1.069
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr13_-_27572646
|
1.066
|
NM_004119
|
FLT3
|
fms-related tyrosine kinase 3
|
|
chr17_-_1029758
|
1.062
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr3_-_53055078
|
1.032
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr6_-_80713589
|
1.030
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr1_-_233880676
|
1.030
|
NM_001098721
NM_004485
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr17_-_78249750
|
1.028
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr4_+_7245218
|
1.024
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr2_-_106869949
|
1.012
|
NM_001142352
NM_032528
|
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2
|
|
chr2_+_176761552
|
1.010
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr5_+_167889123
|
1.008
|
|
|
|
|
chr4_+_3264509
|
1.000
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr16_-_17472238
|
0.998
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr1_+_221633337
|
0.997
|
NM_152610
|
C1orf65
|
chromosome 1 open reading frame 65
|
|
chr22_+_49459935
|
0.996
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr16_-_4528380
|
0.996
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr2_-_128792515
|
0.992
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr2_+_233448739
|
0.984
|
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr21_-_43671355
|
0.983
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr1_-_199704773
|
0.978
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr8_+_120289735
|
0.977
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr6_+_36206239
|
0.973
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr2_+_238265519
|
0.971
|
NM_001137551
NM_001137552
NM_001137553
NM_004735
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr11_-_45643711
|
0.971
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr16_+_11669736
|
0.971
|
NM_003498
|
SNN
|
stannin
|
|
chr19_+_46574277
|
0.967
|
NM_001098821
NM_001098822
NM_001098823
NM_001098824
|
TMEM91
|
transmembrane protein 91
|
|
chr1_+_15957832
|
0.958
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr15_+_49421004
|
0.955
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr3_+_53170254
|
0.955
|
NM_006254
NM_212539
|
PRKCD
|
protein kinase C, delta
|
|
chr15_+_99237555
|
0.928
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr14_+_89597860
|
0.925
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr3_+_120904555
|
0.923
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr1_+_107485071
|
0.920
|
|
NTNG1
|
netrin G1
|
|
chr16_+_87047418
|
0.916
|
NM_153813
|
ZFPM1
|
zinc finger protein, multitype 1
|
|
chr1_-_48235148
|
0.915
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr3_-_126414252
|
0.909
|
NM_024628
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr8_-_101227203
|
0.907
|
NM_001029860
|
FBXO43
|
F-box protein 43
|
|
chr11_+_118544625
|
0.907
|
NM_024618
NM_170722
|
NLRX1
|
NLR family member X1
|
|
chr17_-_43470134
|
0.904
|
NM_016429
|
COPZ2
|
coatomer protein complex, subunit zeta 2
|
|
chr9_-_6635564
|
0.903
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr8_+_25098296
|
0.898
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr18_-_7107757
|
0.890
|
NM_005559
|
LAMA1
|
laminin, alpha 1
|
|
chr20_-_49818306
|
0.885
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr17_-_53387493
|
0.877
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr14_+_103251798
|
0.876
|
NM_001198953
NM_024071
|
ZFYVE21
|
zinc finger, FYVE domain containing 21
|
|
chr1_+_208178198
|
0.874
|
|
SYT14
|
synaptotagmin XIV
|
|
chr19_+_61798434
|
0.874
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr10_+_129595349
|
0.871
|
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr2_+_27163114
|
0.867
|
NM_000221
NM_006488
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr1_+_199127242
|
0.866
|
NM_018265
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr18_+_17076166
|
0.866
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr11_-_64247230
|
0.865
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr14_+_104935897
|
0.862
|
NM_001195082
|
LOC647310
|
testis expressed gene 22
|
|
chr5_+_135392482
|
0.860
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr22_+_48966394
|
0.859
|
|
TRABD
|
TraB domain containing
|
|
chr15_+_57517564
|
0.858
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr7_-_150847884
|
0.853
|
NM_005614
|
RHEB
|
Ras homolog enriched in brain
|
|
chr7_-_117300702
|
0.853
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr6_+_17389752
|
0.852
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_+_235525355
|
0.848
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr11_+_7997366
|
0.847
|
|
TUB
|
tubby homolog (mouse)
|
|
chr17_+_77780177
|
0.841
|
NM_004207
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr1_-_223907283
|
0.840
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr3_+_9826930
|
0.840
|
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3
|
|
chr14_+_102128844
|
0.839
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr12_+_13044604
|
0.838
|
|
HTR7P1
|
5-hydroxytryptamine (serotonin) receptor 7 pseudogene 1
|
|
chr20_-_61933012
|
0.837
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr8_+_25098171
|
0.836
|
NM_024940
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr17_-_25112393
|
0.835
|
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr14_+_20608179
|
0.832
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr10_-_71576335
|
0.830
|
|
|
|
|
chr1_-_6468108
|
0.829
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr1_-_19998955
|
0.829
|
NM_181719
|
TMCO4
|
transmembrane and coiled-coil domains 4
|
|
chr13_+_35904408
|
0.827
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr4_-_83702123
|
0.827
|
NM_001080506
|
TMEM150C
|
transmembrane protein 150C
|
|
chr5_-_114533448
|
0.826
|
|
TRIM36
|
tripartite motif containing 36
|
|
chr22_-_26527469
|
0.823
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr17_-_1412774
|
0.823
|
NM_006224
|
PITPNA
|
phosphatidylinositol transfer protein, alpha
|
|
chr1_+_1971762
|
0.822
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr3_-_127558747
|
0.816
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr20_+_8060762
|
0.816
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr17_+_77779570
|
0.816
|
NM_001042422
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr8_+_136538739
|
0.815
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr1_-_6243621
|
0.814
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr5_+_68823874
|
0.807
|
NM_002538
|
OCLN
|
occludin
|
|
chr5_-_58370693
|
0.798
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_+_22171150
|
0.795
|
NM_001142649
NM_213599
|
ANO5
|
anoctamin 5
|
|
chr20_-_49818268
|
0.795
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr4_+_995609
|
0.787
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr10_-_131652364
|
0.780
|
|
EBF3
|
early B-cell factor 3
|
|
chr4_-_170428681
|
0.779
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr3_-_51976428
|
0.779
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr4_+_72271218
|
0.776
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr2_+_9532087
|
0.776
|
NM_001039613
|
IAH1
|
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae)
|
|
chr10_-_135036805
|
0.776
|
NM_004092
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial
|
|
chr17_+_78630793
|
0.775
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr17_-_24068759
|
0.769
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr3_-_185461768
|
0.767
|
NM_033259
|
CAMK2N2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2
|
|
chr2_+_242289983
|
0.767
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr2_-_20288647
|
0.766
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr9_-_139068269
|
0.762
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr7_+_1536847
|
0.761
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr10_+_134972902
|
0.759
|
|
|
|
|
chr6_-_10527772
|
0.756
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr2_-_128149112
|
0.755
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr15_-_58671929
|
0.747
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_+_92575767
|
0.747
|
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr22_-_39964001
|
0.745
|
|
CHADL
|
chondroadherin-like
|
|
chr2_-_42574587
|
0.745
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr4_-_170428564
|
0.744
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chrX_-_1531733
|
0.744
|
NM_001173474
NM_004192
|
ASMTL
|
acetylserotonin O-methyltransferase-like
|
|
chr8_-_37876129
|
0.741
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr11_+_120828058
|
0.738
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr17_+_43165608
|
0.738
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr7_+_55054152
|
0.737
|
NM_005228
NM_201282
NM_201283
NM_201284
|
EGFR
|
epidermal growth factor receptor
|
|
chr3_+_54131731
|
0.735
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr16_-_66258072
|
0.731
|
NM_032140
|
C16orf48
|
chromosome 16 open reading frame 48
|
|
chr16_-_11588525
|
0.731
|
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr12_-_105165805
|
0.730
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr1_-_85131439
|
0.728
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr11_-_19219720
|
0.725
|
|
E2F8
|
E2F transcription factor 8
|
|
chr3_-_198180005
|
0.725
|
NM_025163
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z
|
|
chr16_-_65742323
|
0.723
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr2_-_135193011
|
0.722
|
NM_030923
|
TMEM163
|
transmembrane protein 163
|
|
chr5_-_168660293
|
0.713
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr21_+_17807167
|
0.712
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|