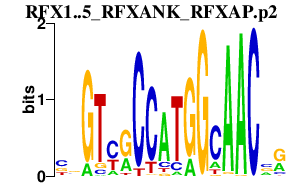
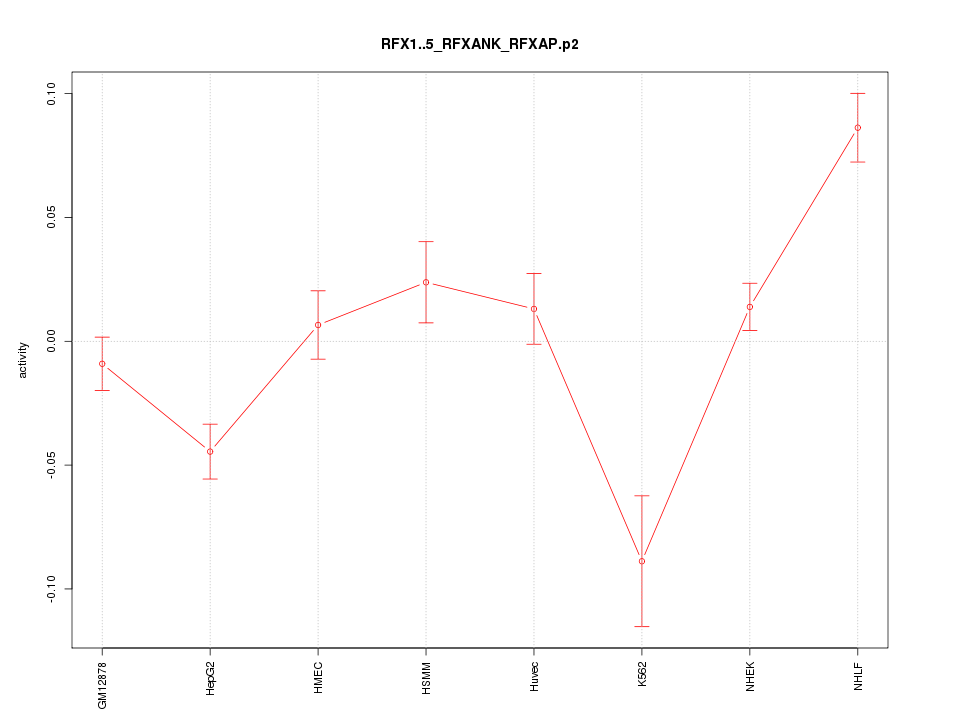
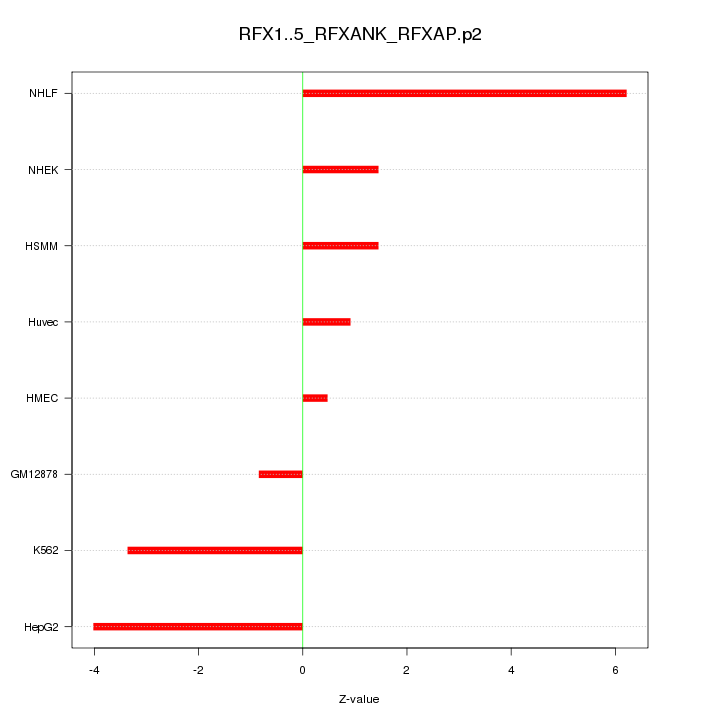
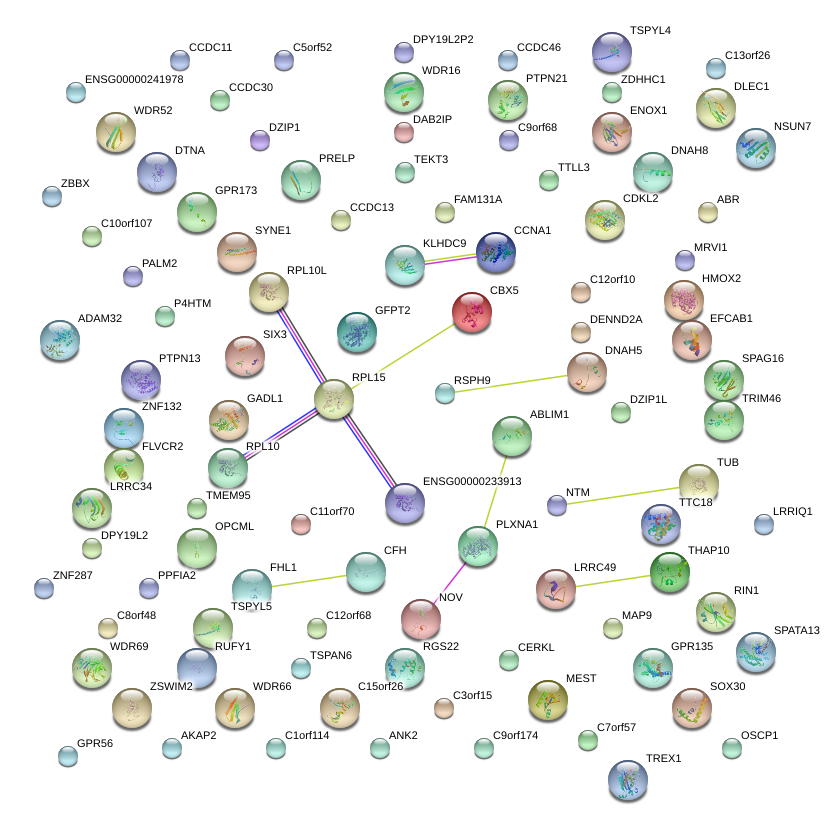
|
chr1_+_42701460
|
10.271
|
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr2_-_187422141
|
7.732
|
NM_182521
|
ZSWIM2
|
zinc finger, SWIM-type containing 2
|
|
chr5_-_157031022
|
7.527
|
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr5_+_157031138
|
6.193
|
NM_001145132
|
C5orf52
|
chromosome 5 open reading frame 52
|
|
chr4_-_156517423
|
5.837
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr18_+_30544193
|
5.314
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_-_156517394
|
5.172
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr19_-_63643400
|
5.113
|
NM_003433
|
ZNF132
|
zinc finger protein 132
|
|
chr12_+_46863632
|
5.090
|
NM_001013635
|
C12orf68
|
chromosome 12 open reading frame 68
|
|
chr7_+_48041632
|
4.995
|
NM_001100159
|
C7orf57
|
chromosome 7 open reading frame 57
|
|
chr8_+_120497732
|
4.779
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr12_-_62348506
|
4.779
|
NM_173812
|
DPY19L2
|
dpy-19-like 2 (C. elegans)
|
|
chr4_-_76774594
|
4.761
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr18_+_76006795
|
4.549
|
|
LOC100130522
|
hypothetical LOC100130522
|
|
chr4_-_156517544
|
4.516
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chr11_+_7997366
|
4.344
|
|
TUB
|
tubby homolog (mouse)
|
|
chr1_+_159334774
|
4.294
|
NM_001007255
NM_152366
|
KLHDC9
|
kelch domain containing 9
|
|
chr3_+_120904555
|
4.247
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr13_-_95092283
|
4.225
|
|
DZIP1
|
DAZ interacting protein 1
|
|
chr3_-_114643008
|
4.186
|
NM_001164496
NM_018338
|
WDR52
|
WD repeat domain 52
|
|
chr17_-_16413188
|
4.138
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr8_+_13468722
|
4.113
|
NM_001007090
|
C8orf48
|
chromosome 8 open reading frame 48
|
|
chr14_-_88087575
|
4.070
|
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr9_+_111442888
|
4.055
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr15_-_68971665
|
3.862
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr6_+_43720744
|
3.780
|
NM_001193341
NM_152732
|
RSPH9
|
radial spoke head 9 homolog (Chlamydomonas)
|
|
chr1_-_167663278
|
3.754
|
NM_021179
|
C1orf114
|
chromosome 1 open reading frame 114
|
|
chr3_-_171013000
|
3.741
|
|
LRRC34
|
leucine rich repeat containing 34
|
|
chr1_-_36688556
|
3.705
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr15_+_68971835
|
3.693
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr10_-_74788508
|
3.602
|
NM_145170
|
TTC18
|
tetratricopeptide repeat domain 18
|
|
chr6_-_152744143
|
3.583
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_+_99109730
|
3.556
|
NM_020893
|
C9orf174
|
chromosome 9 open reading frame 174
|
|
chr8_-_49810270
|
3.515
|
NM_001142857
NM_024593
|
EFCAB1
|
EF-hand calcium binding domain 1
|
|
chr3_+_9826643
|
3.502
|
NM_001025930
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3
|
|
chr3_-_139315011
|
3.494
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr7_-_139987044
|
3.452
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr17_-_959007
|
3.393
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr13_-_43259032
|
3.361
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr12_+_51979721
|
3.356
|
NM_021640
|
C12orf10
|
chromosome 12 open reading frame 10
|
|
chr9_+_123501186
|
3.350
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr1_+_153413675
|
3.327
|
|
TRIM46
|
tripartite motif containing 46
|
|
chr10_+_63092724
|
3.317
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr6_-_152744022
|
3.310
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr11_+_131285747
|
3.263
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr11_+_101423378
|
3.233
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr13_+_23451919
|
3.222
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr7_-_35192177
|
3.208
|
|
DPY19L2P1
|
dpy-19-like 2 pseudogene 1 (C. elegans)
|
|
chr4_+_114433551
|
3.106
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr13_+_30404809
|
3.079
|
NM_152325
|
C13orf26
|
chromosome 13 open reading frame 26
|
|
chr12_+_120840829
|
3.071
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr4_+_87734489
|
3.002
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr5_+_178919298
|
2.980
|
NM_001040451
NM_001040452
|
RUFY1
|
RUN and FYVE domain containing 1
|
|
chr5_-_14064846
|
2.922
|
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr3_+_9826930
|
2.921
|
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3
|
|
chr17_-_5345167
|
2.915
|
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr2_+_228444570
|
2.912
|
NM_178821
|
WDR69
|
WD repeat domain 69
|
|
chr12_-_52939636
|
2.910
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chrX_+_53095230
|
2.897
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr17_-_5344898
|
2.892
|
NM_001162371
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr2_-_182229977
|
2.832
|
NM_001030311
NM_001030312
NM_001030313
NM_001160277
NM_201548
|
CERKL
|
ceramide kinase-like
|
|
chr18_-_46046771
|
2.803
|
NM_145020
|
CCDC11
|
coiled-coil domain containing 11
|
|
chr3_-_30911156
|
2.802
|
NM_207359
|
GADL1
|
glutamate decarboxylase-like 1
|
|
chr17_-_15185623
|
2.787
|
NM_031898
|
TEKT3
|
tektin 3
|
|
chr3_+_49002774
|
2.744
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr17_+_17339059
|
2.741
|
|
|
|
|
chr15_+_79213660
|
2.701
|
NM_173528
|
C15orf26
|
chromosome 15 open reading frame 26
|
|
chr13_+_35904408
|
2.671
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr12_-_80676562
|
2.648
|
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr3_-_42789738
|
2.610
|
NM_144719
|
CCDC13
|
coiled-coil domain containing 13
|
|
chr2_+_45022540
|
2.591
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chr8_-_98359248
|
2.570
|
|
TSPYL5
|
TSPY-like 5
|
|
chr3_-_168580764
|
2.540
|
NM_024687
|
ZBBX
|
zinc finger, B-box domain containing
|
|
chr10_-_116382089
|
2.539
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_+_38055699
|
2.533
|
NM_007335
NM_007337
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chr8_-_101187346
|
2.529
|
|
RGS22
|
regulator of G-protein signaling 22
|
|
chr17_+_7199178
|
2.517
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chrX_+_135079461
|
2.494
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr3_-_171013145
|
2.481
|
NM_001172779
NM_001172780
NM_153353
|
LRRC34
|
leucine rich repeat containing 34
|
|
chr7_-_102707901
|
2.476
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr11_-_65860450
|
2.452
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr8_+_39084206
|
2.417
|
NM_145004
|
ADAM32
|
ADAM metallopeptidase domain 32
|
|
chr5_-_179712920
|
2.376
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr12_+_83954229
|
2.366
|
NM_001079910
NM_032165
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr16_+_56219802
|
2.352
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr11_-_132318741
|
2.343
|
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr7_+_129919330
|
2.329
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr3_+_185538998
|
2.307
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr1_+_149960636
|
2.301
|
NM_001144956
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1
|
|
chr8_-_98359347
|
2.270
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr7_+_129919157
|
2.263
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_129919134
|
2.233
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr8_-_98359366
|
2.217
|
|
TSPYL5
|
TSPY-like 5
|
|
chr11_-_10671873
|
2.196
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr16_-_66007650
|
2.194
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr4_+_40446670
|
2.187
|
NM_024677
|
NSUN7
|
NOP2/Sun domain family, member 7
|
|
chr17_+_9420668
|
2.187
|
NM_001080556
NM_145054
|
WDR16
|
WD repeat domain 16
|
|
chrX_-_99778340
|
2.182
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr17_-_61618276
|
2.180
|
NM_145036
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr1_+_201711505
|
2.177
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr9_-_4656665
|
2.173
|
NM_001039395
|
C9orf68
|
chromosome 9 open reading frame 68
|
|
chr2_+_213857347
|
2.169
|
NM_001025436
NM_024532
|
SPAG16
|
sperm associated antigen 16
|
|
chr14_-_59001603
|
2.161
|
|
GPR135
|
G protein-coupled receptor 135
|
|
chr3_+_48482213
|
2.155
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr14_+_75114692
|
2.153
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr16_+_4485859
|
2.130
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr10_+_105243724
|
2.129
|
NM_004210
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr12_-_52939584
|
2.128
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr12_-_83954185
|
2.120
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr1_-_215870935
|
2.116
|
NM_018040
|
GPATCH2
|
G patch domain containing 2
|
|
chr1_-_51660328
|
2.088
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr4_-_148086413
|
2.086
|
NM_031956
|
TTC29
|
tetratricopeptide repeat domain 29
|
|
chr3_-_50357994
|
2.081
|
NM_015896
|
ZMYND10
|
zinc finger, MYND-type containing 10
|
|
chr1_+_201711578
|
2.078
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr1_+_15446354
|
2.076
|
NM_052929
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1
|
|
chr14_+_99329198
|
2.055
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr17_+_19255083
|
2.032
|
NM_007148
|
RNF112
|
ring finger protein 112
|
|
chr11_+_6998275
|
2.026
|
NM_176822
|
NLRP14
|
NLR family, pyrin domain containing 14
|
|
chr12_+_79362256
|
2.000
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr11_-_6998041
|
1.997
|
NM_013249
|
ZNF214
|
zinc finger protein 214
|
|
chr11_-_75057062
|
1.981
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chr22_-_42539399
|
1.981
|
NM_022785
NM_198856
|
EFCAB6
|
EF-hand calcium binding domain 6
|
|
chr12_-_88270375
|
1.975
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr12_+_50104936
|
1.970
|
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr14_-_59001804
|
1.961
|
NM_022571
|
GPR135
|
G protein-coupled receptor 135
|
|
chr11_-_10671736
|
1.947
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr5_-_16795336
|
1.935
|
|
MYO10
|
myosin X
|
|
chr1_+_84402962
|
1.919
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chrX_+_135079120
|
1.900
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_-_10671608
|
1.896
|
NM_001100167
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr3_-_27385856
|
1.891
|
NM_199347
|
NEK10
|
NIMA (never in mitosis gene a)- related kinase 10
|
|
chr12_-_123568781
|
1.885
|
|
|
|
|
chr12_+_50104859
|
1.867
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr14_-_59407103
|
1.858
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr8_-_101187519
|
1.825
|
NM_015668
|
RGS22
|
regulator of G-protein signaling 22
|
|
chr1_-_86634511
|
1.817
|
NM_001007022
NM_001184765
NM_001184766
NM_020729
|
ODF2L
|
outer dense fiber of sperm tails 2-like
|
|
chr12_-_88270369
|
1.790
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr11_+_85763425
|
1.774
|
NM_001156474
NM_021827
|
CCDC81
|
coiled-coil domain containing 81
|
|
chr7_+_21549357
|
1.774
|
NM_003777
|
DNAH11
|
dynein, axonemal, heavy chain 11
|
|
chr22_-_43272567
|
1.753
|
NM_032287
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr19_+_63237211
|
1.739
|
NM_182572
|
ZSCAN1
|
zinc finger and SCAN domain containing 1
|
|
chr11_-_114880275
|
1.738
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr15_-_88034857
|
1.732
|
NM_003847
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha
|
|
chr11_+_26310253
|
1.729
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr17_+_45940911
|
1.722
|
|
MYCBPAP
|
MYCBP associated protein
|
|
chr5_-_142045770
|
1.719
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chrX_+_103243762
|
1.718
|
NM_001143978
|
ZCCHC18
|
zinc finger, CCHC domain containing 18
|
|
chr18_+_42780784
|
1.716
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr21_-_45532176
|
1.715
|
NM_015227
NM_133635
|
POFUT2
|
protein O-fucosyltransferase 2
|
|
chr14_+_23908141
|
1.713
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr19_-_63301402
|
1.705
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr5_+_92944680
|
1.700
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr12_+_104248769
|
1.674
|
|
C12orf75
|
chromosome 12 open reading frame 75
|
|
chr13_+_23451838
|
1.667
|
|
|
|
|
chr18_-_45630154
|
1.618
|
|
MYO5B
|
myosin VB
|
|
chr7_-_102708076
|
1.612
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr19_-_51690989
|
1.596
|
NM_020709
|
PNMAL2
|
PNMA-like 2
|
|
chr19_-_63301418
|
1.591
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr15_+_88035031
|
1.586
|
NM_020212
|
WDR93
|
WD repeat domain 93
|
|
chr9_-_109291866
|
1.584
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr16_-_674318
|
1.577
|
|
JMJD8
|
jumonji domain containing 8
|
|
chr8_-_485330
|
1.571
|
NM_175075
|
C8orf42
|
chromosome 8 open reading frame 42
|
|
chr19_-_63301386
|
1.570
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr1_-_48710263
|
1.567
|
|
SPATA6
|
spermatogenesis associated 6
|
|
chr22_+_21817512
|
1.559
|
NM_004914
|
RAB36
|
RAB36, member RAS oncogene family
|
|
chr18_+_50049846
|
1.551
|
NM_007195
|
POLI
|
polymerase (DNA directed) iota
|
|
chr13_-_59223074
|
1.540
|
|
|
|
|
chr6_+_117044334
|
1.540
|
NM_001010892
NM_001161664
|
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas)
|
|
chr17_-_71648965
|
1.537
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr8_-_94781836
|
1.536
|
|
LOC642924
|
hypothetical LOC642924
|
|
chr19_-_584526
|
1.535
|
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr9_+_115396020
|
1.526
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr19_-_584563
|
1.518
|
NM_005035
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr22_+_25383392
|
1.517
|
|
MIAT
|
myocardial infarction associated transcript (non-protein coding)
|
|
chr11_+_85243791
|
1.511
|
NM_173556
|
CCDC83
|
coiled-coil domain containing 83
|
|
chr19_-_7459867
|
1.511
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr11_+_133444175
|
1.504
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr19_-_584474
|
1.501
|
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr1_+_215871288
|
1.499
|
NM_138796
|
SPATA17
|
spermatogenesis associated 17
|
|
chr16_-_674357
|
1.492
|
NM_001005920
|
JMJD8
|
jumonji domain containing 8
|
|
chr2_-_233501069
|
1.459
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr5_+_43228083
|
1.458
|
NM_153361
|
MGC42105
|
serine/threonine-protein kinase NIM1
|
|
chr1_+_33319287
|
1.447
|
NM_052998
|
ADC
|
arginine decarboxylase
|
|
chr6_-_165643064
|
1.435
|
NM_144980
|
C6orf118
|
chromosome 6 open reading frame 118
|
|
chr11_+_120399966
|
1.429
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chrX_-_71268475
|
1.427
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr7_+_156126157
|
1.426
|
NM_030936
NM_001184996
|
RNF32
|
ring finger protein 32
|
|
chr11_+_93885342
|
1.421
|
NM_001190462
|
LOC643037
|
hypothetical protein LOC643037
|
|
chr9_-_129537372
|
1.410
|
NM_001085347
NM_001134430
NM_001134431
NM_130459
|
TOR2A
|
torsin family 2, member A
|
|
chr3_+_50281582
|
1.399
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr5_-_54565181
|
1.396
|
NM_021147
|
CCNO
|
cyclin O
|
|
chr8_+_94781910
|
1.394
|
NM_145269
|
FAM92A1
|
family with sequence similarity 92, member A1
|
|
chr3_-_58538110
|
1.383
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr3_+_156280129
|
1.372
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_+_37795101
|
1.362
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr14_+_105063975
|
1.349
|
NM_025268
|
TMEM121
|
transmembrane protein 121
|
|
chr11_-_56846246
|
1.344
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr4_-_87500201
|
1.340
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr1_-_48710378
|
1.336
|
NM_019073
|
SPATA6
|
spermatogenesis associated 6
|
|
chr22_-_16887151
|
1.334
|
NM_001122731
NM_001136004
NM_015241
|
MICAL3
|
microtubule associated monoxygenase, calponin and LIM domain containing 3
|