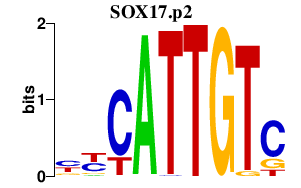
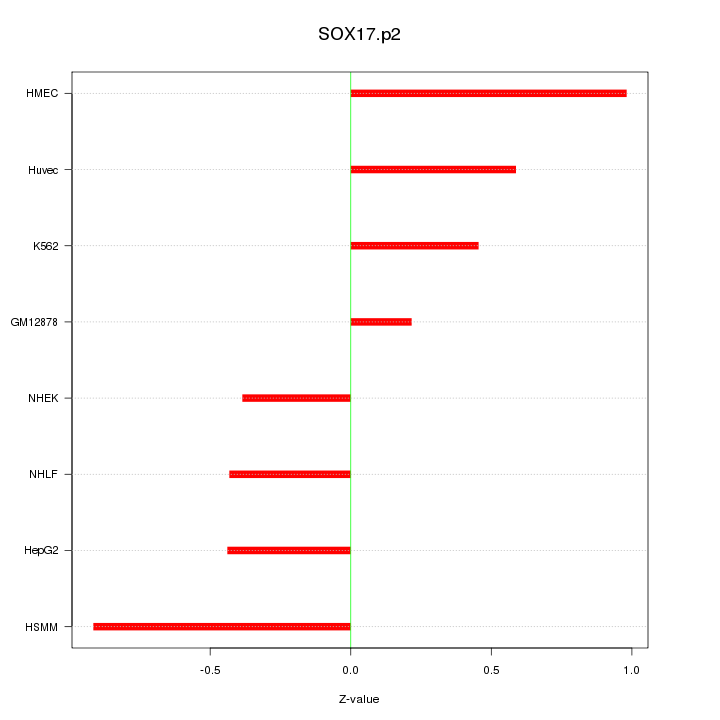
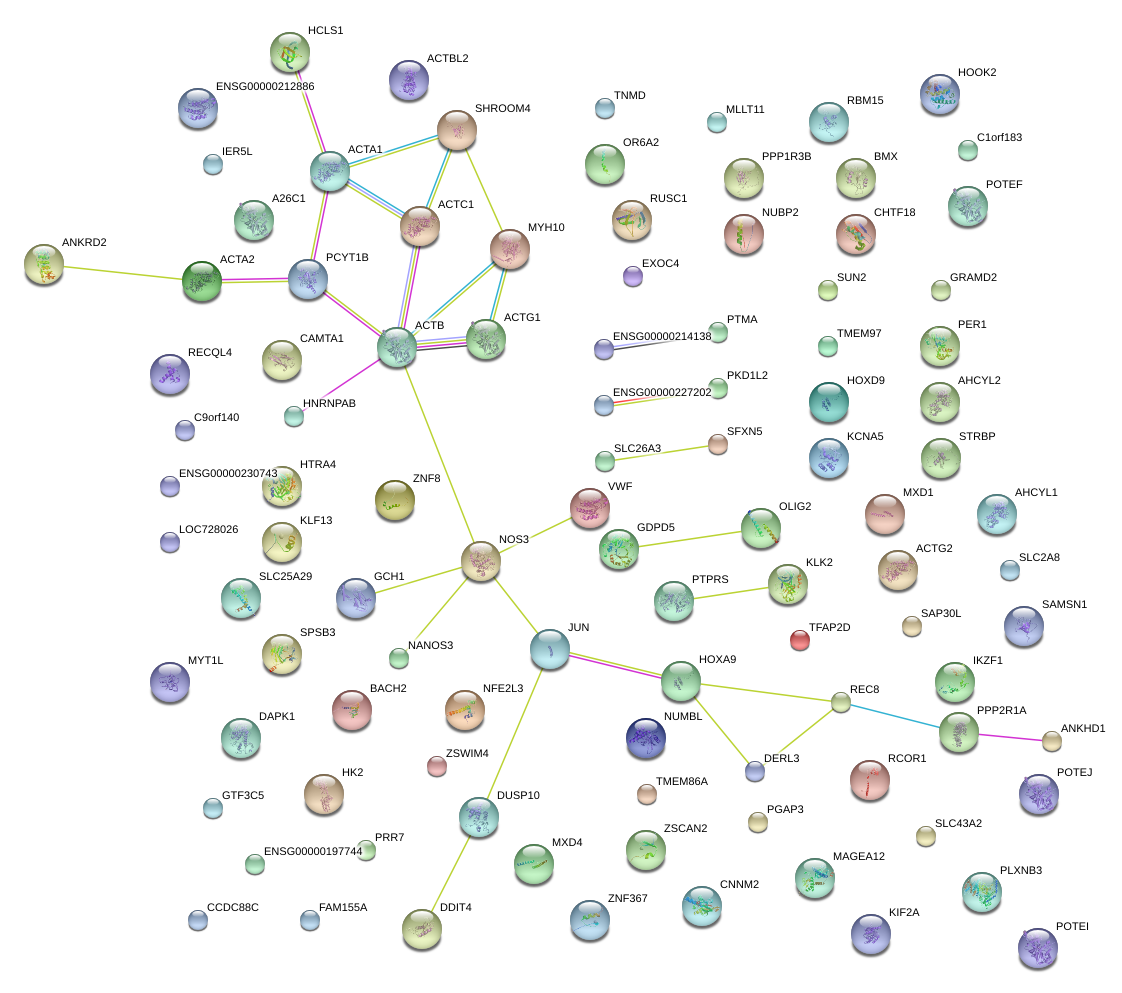
|
chr7_+_128802719
|
0.698
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr4_-_2233606
|
0.672
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr14_+_23711044
|
0.654
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr15_+_29406335
|
0.646
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr17_-_59817654
|
0.642
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_-_59817575
|
0.621
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_-_59817456
|
0.597
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_-_59817722
|
0.595
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr5_+_177564140
|
0.555
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr2_+_74914742
|
0.528
|
|
HK2
|
hexokinase 2
|
|
chr22_+_22703107
|
0.517
|
NM_001144931
|
LOC391322
|
D-dopachrome tautomerase-like
|
|
chr1_+_6767968
|
0.506
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr16_-_1772305
|
0.501
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr14_+_23710924
|
0.492
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr17_-_8474733
|
0.484
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr11_-_74914514
|
0.478
|
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr7_+_150321771
|
0.477
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr13_-_107317460
|
0.472
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr17_-_59818033
|
0.466
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr16_-_1772572
|
0.465
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr5_+_177564139
|
0.461
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr16_-_1772577
|
0.457
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr19_+_63482118
|
0.454
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chrX_+_152684150
|
0.441
|
|
PLXNB3
|
plexin B3
|
|
chr14_-_99842519
|
0.441
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr17_+_23670247
|
0.440
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr1_+_110683586
|
0.431
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr19_+_47593137
|
0.431
|
|
|
|
|
chrX_-_50403412
|
0.431
|
|
SHROOM4
|
shroom family member 4
|
|
chr5_+_176807931
|
0.422
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr5_+_177564102
|
0.411
|
NM_004499
NM_031266
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr2_+_232281491
|
0.407
|
|
PTMA
|
prothymosin, alpha
|
|
chr17_-_1478293
|
0.407
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr9_-_130980302
|
0.406
|
|
IER5L
|
immediate early response 5-like
|
|
chr9_-_130980359
|
0.405
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr9_+_129199204
|
0.399
|
NM_014580
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8
|
|
chrX_+_15428820
|
0.397
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr19_-_45888259
|
0.388
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr6_-_91063181
|
0.388
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr4_-_2233504
|
0.387
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr1_-_153560532
|
0.387
|
NM_001039517
|
C1orf104
|
chromosome 1 open reading frame 104
|
|
chr2_+_69995740
|
0.383
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr7_+_50314801
|
0.382
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr1_+_110683580
|
0.381
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr10_+_104667991
|
0.381
|
NM_017649
NM_199076
NM_199077
|
CNNM2
|
cyclin M2
|
|
chr8_-_9046460
|
0.374
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr7_-_27171648
|
0.373
|
|
HOXA9
|
homeobox A9
|
|
chr14_-_54438913
|
0.368
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr7_+_148613304
|
0.365
|
|
LOC155060
|
AI894139 pseudogene
|
|
chrX_+_48934831
|
0.365
|
|
|
|
|
chr12_-_6103945
|
0.363
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr14_+_102128741
|
0.363
|
|
RCOR1
|
REST corepressor 1
|
|
chr9_-_98220482
|
0.362
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr9_-_125070613
|
0.361
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr7_+_26158257
|
0.361
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr11_-_6773589
|
0.359
|
NM_003696
|
OR6A2
|
olfactory receptor, family 6, subfamily A, member 2
|
|
chr14_-_90953883
|
0.357
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr9_-_139084833
|
0.354
|
NM_178448
|
C9orf140
|
chromosome 9 open reading frame 140
|
|
chr17_+_37373195
|
0.351
|
|
|
|
|
chr2_-_2313894
|
0.350
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr7_-_107230867
|
0.348
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr1_-_112083387
|
0.346
|
NM_019099
|
C1orf183
|
chromosome 1 open reading frame 183
|
|
chr1_+_110683467
|
0.345
|
NM_022768
|
RBM15
|
RNA binding motif protein 15
|
|
chr7_-_27171600
|
0.343
|
|
HOXA9
|
homeobox A9
|
|
chr8_+_38950824
|
0.343
|
NM_153692
|
HTRA4
|
HtrA serine peptidase 4
|
|
chr6_+_50789215
|
0.341
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr10_+_73703682
|
0.341
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr19_+_57385146
|
0.341
|
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr9_+_134895882
|
0.339
|
NM_001122823
NM_012087
|
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa
|
|
chr21_-_14840506
|
0.338
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr1_+_149299530
|
0.337
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr22_-_37481925
|
0.337
|
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr21_+_33320080
|
0.337
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr15_+_82945223
|
0.336
|
NM_001007072
NM_017894
NM_181877
|
ZSCAN2
|
zinc finger and SCAN domain containing 2
|
|
chr19_-_5291700
|
0.332
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr2_-_73151894
|
0.331
|
|
SFXN5
|
sideroflexin 5
|
|
chr2_+_69995656
|
0.328
|
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr5_+_153805761
|
0.327
|
|
SAP30L
|
SAP30-like
|
|
chrX_-_24600661
|
0.327
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr19_+_57385130
|
0.323
|
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr9_-_98220360
|
0.322
|
|
ZNF367
|
zinc finger protein 367
|
|
chr7_-_27171673
|
0.321
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr1_+_149299490
|
0.320
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr16_+_1772929
|
0.319
|
NM_012225
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr22_-_22511173
|
0.319
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr1_+_153560841
|
0.314
|
NM_014328
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr11_+_18676857
|
0.314
|
|
TMEM86A
|
transmembrane protein 86A
|
|
chr9_+_89302574
|
0.312
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr15_-_70277163
|
0.312
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr19_+_13767207
|
0.309
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr16_+_778646
|
0.309
|
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr7_-_5535432
|
0.307
|
|
ACTB
|
actin, beta
|
|
chr17_-_7996409
|
0.301
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr19_+_57385140
|
0.301
|
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr17_-_77093603
|
0.297
|
|
ACTG1
|
actin, gamma 1
|
|
chr3_-_122862447
|
0.297
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr16_-_79811475
|
0.296
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr6_-_91063301
|
0.295
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr8_-_145713959
|
0.293
|
NM_004260
|
RECQL4
|
RecQ protein-like 4
|
|
chr5_+_139761579
|
0.293
|
NM_020690
NM_001197030
NM_017747
NM_017978
NM_024668
|
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough
ankyrin repeat and KH domain containing 1
|
|
chr1_-_59022275
|
0.293
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chrX_+_99726445
|
0.291
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr1_-_219982081
|
0.290
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_+_1971762
|
0.290
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr7_+_882704
|
0.287
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr22_-_48607449
|
0.286
|
|
BRD1
|
bromodomain containing 1
|
|
chr9_-_125070675
|
0.286
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr11_+_18676913
|
0.284
|
NM_153347
|
TMEM86A
|
transmembrane protein 86A
|
|
chr3_-_122862391
|
0.283
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr1_+_165564904
|
0.283
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_+_152608159
|
0.283
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr13_-_40138607
|
0.282
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr17_+_43373887
|
0.282
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr1_+_149299504
|
0.281
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr15_-_38188333
|
0.280
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr22_+_42900168
|
0.280
|
NM_001137605
|
PARVG
|
parvin, gamma
|
|
chr2_+_12775814
|
0.280
|
|
|
|
|
chr10_+_70748581
|
0.279
|
NM_000188
|
HK1
|
hexokinase 1
|
|
chr16_-_1601970
|
0.279
|
NM_014714
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas)
|
|
chr17_+_72376022
|
0.278
|
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr10_+_3099708
|
0.278
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr2_-_157890400
|
0.277
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr7_+_882766
|
0.276
|
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr18_+_3441589
|
0.275
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr19_+_61624
|
0.274
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr8_+_95977135
|
0.273
|
|
C8orf38
|
chromosome 8 open reading frame 38
|
|
chr9_+_80101810
|
0.272
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr19_+_60543032
|
0.271
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr17_+_71095733
|
0.271
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr5_+_78568094
|
0.271
|
|
|
|
|
chr17_-_59131210
|
0.270
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr19_-_48791967
|
0.270
|
NM_001007561
|
IRGQ
|
immunity-related GTPase family, Q
|
|
chr17_-_44058612
|
0.266
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr13_+_45174446
|
0.266
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chr7_-_5429702
|
0.264
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr1_+_165565006
|
0.264
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr17_-_43288137
|
0.264
|
NM_199262
|
SP6
|
Sp6 transcription factor
|
|
chr1_-_202730135
|
0.262
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr9_-_139316523
|
0.262
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr7_-_5535726
|
0.261
|
|
ACTB
|
actin, beta
|
|
chr5_+_102493155
|
0.261
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr19_+_40824486
|
0.261
|
NM_014209
|
ETV2
|
ets variant 2
|
|
chr11_+_46358909
|
0.259
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr16_+_55523526
|
0.258
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr4_-_41445479
|
0.256
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr11_-_16380967
|
0.254
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr19_-_8314120
|
0.254
|
NM_198471
|
KANK3
|
KN motif and ankyrin repeat domains 3
|
|
chr19_+_57385072
|
0.252
|
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr7_-_5535349
|
0.251
|
|
ACTB
|
actin, beta
|
|
chr1_-_219982016
|
0.250
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr17_+_73511843
|
0.249
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr11_-_2863536
|
0.249
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr10_+_17311242
|
0.247
|
|
VIM
|
vimentin
|
|
chr18_-_75812640
|
0.247
|
NM_001146343
NM_001146345
NM_025078
|
PQLC1
|
PQ loop repeat containing 1
|
|
chr22_-_17892859
|
0.245
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr10_-_99084417
|
0.244
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr1_-_6468108
|
0.243
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr6_+_25862905
|
0.243
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chr21_-_30460841
|
0.242
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr22_-_37481857
|
0.241
|
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr1_+_153560351
|
0.240
|
NM_001105205
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr16_-_4724154
|
0.240
|
NM_133450
|
ANKS3
|
ankyrin repeat and sterile alpha motif domain containing 3
|
|
chr17_-_37590910
|
0.240
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr5_+_153805690
|
0.239
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr21_-_31853160
|
0.238
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr4_-_176970895
|
0.237
|
|
GPM6A
|
glycoprotein M6A
|
|
chr19_+_12910428
|
0.237
|
|
CALR
|
calreticulin
|
|
chr22_+_34126154
|
0.236
|
|
MCM5
|
minichromosome maintenance complex component 5
|
|
chr18_-_22382497
|
0.235
|
NM_001142730
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr6_-_139737042
|
0.235
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr3_-_52287695
|
0.235
|
NM_025222
|
WDR82
|
WD repeat domain 82
|
|
chr10_+_17311282
|
0.234
|
|
VIM
|
vimentin
|
|
chr3_-_52287603
|
0.233
|
|
WDR82
|
WD repeat domain 82
|
|
chr22_-_17799177
|
0.233
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr3_+_48482625
|
0.233
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr3_+_42519098
|
0.233
|
NM_004624
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr1_-_219982049
|
0.232
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_-_38285033
|
0.231
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr6_+_100161370
|
0.230
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr3_+_45610357
|
0.230
|
|
LIMD1
|
LIM domains containing 1
|
|
chr13_-_45654292
|
0.229
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr9_+_139255559
|
0.229
|
|
TUBB2C
|
tubulin, beta 2C
|
|
chr16_-_30010705
|
0.229
|
NM_004608
|
TBX6
|
T-box 6
|
|
chr7_+_143683365
|
0.228
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr14_-_22458220
|
0.228
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr17_-_77093468
|
0.228
|
|
ACTG1
|
actin, gamma 1
|
|
chr13_-_45654296
|
0.227
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_199975674
|
0.225
|
|
NAV1
|
neuron navigator 1
|
|
chr3_+_42519135
|
0.225
|
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr21_-_36870736
|
0.225
|
NM_001146077
|
CLDN14
|
claudin 14
|
|
chr16_+_778635
|
0.224
|
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr19_+_19357629
|
0.223
|
NM_017660
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chr1_-_40140126
|
0.223
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr16_+_28869628
|
0.223
|
NM_032815
|
NFATC2IP
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein
|
|
chr1_-_5975068
|
0.223
|
NM_015102
|
NPHP4
|
nephronophthisis 4
|
|
chr19_-_52243721
|
0.222
|
NM_017854
|
TMEM160
|
transmembrane protein 160
|
|
chr4_-_40553838
|
0.222
|
NM_001166051
NM_001166052
NM_001166053
NM_001166054
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr17_-_77649393
|
0.221
|
NM_004104
|
FASN
|
fatty acid synthase
|
|
chr22_+_48740143
|
0.221
|
NM_001001852
|
PIM3
|
pim-3 oncogene
|
|
chr22_+_34126104
|
0.221
|
NM_006739
|
MCM5
|
minichromosome maintenance complex component 5
|