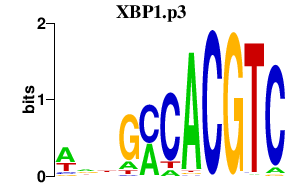
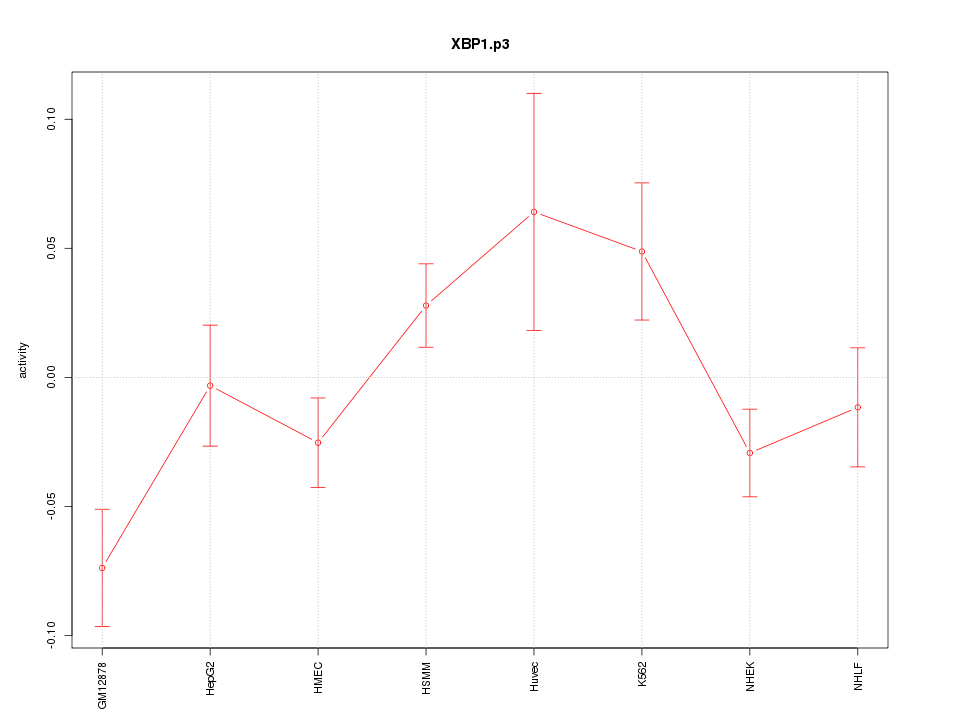
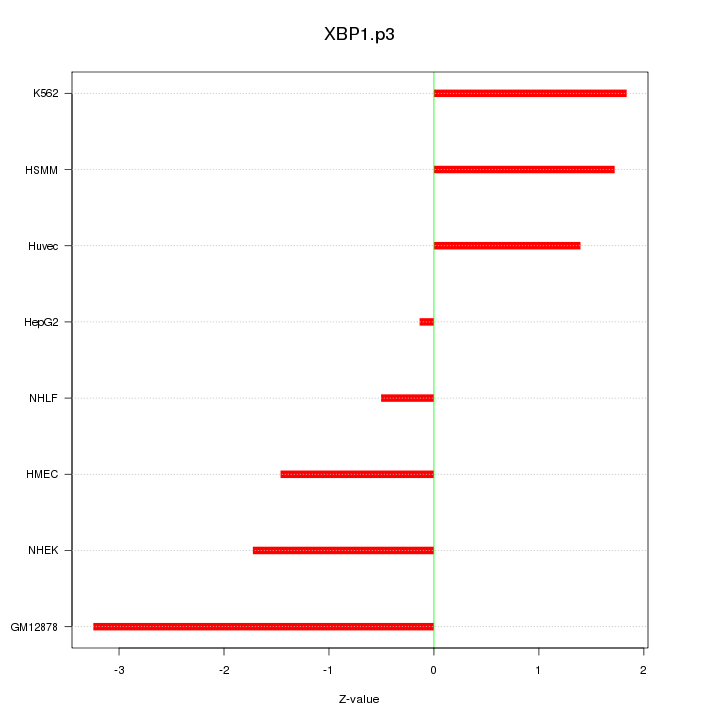
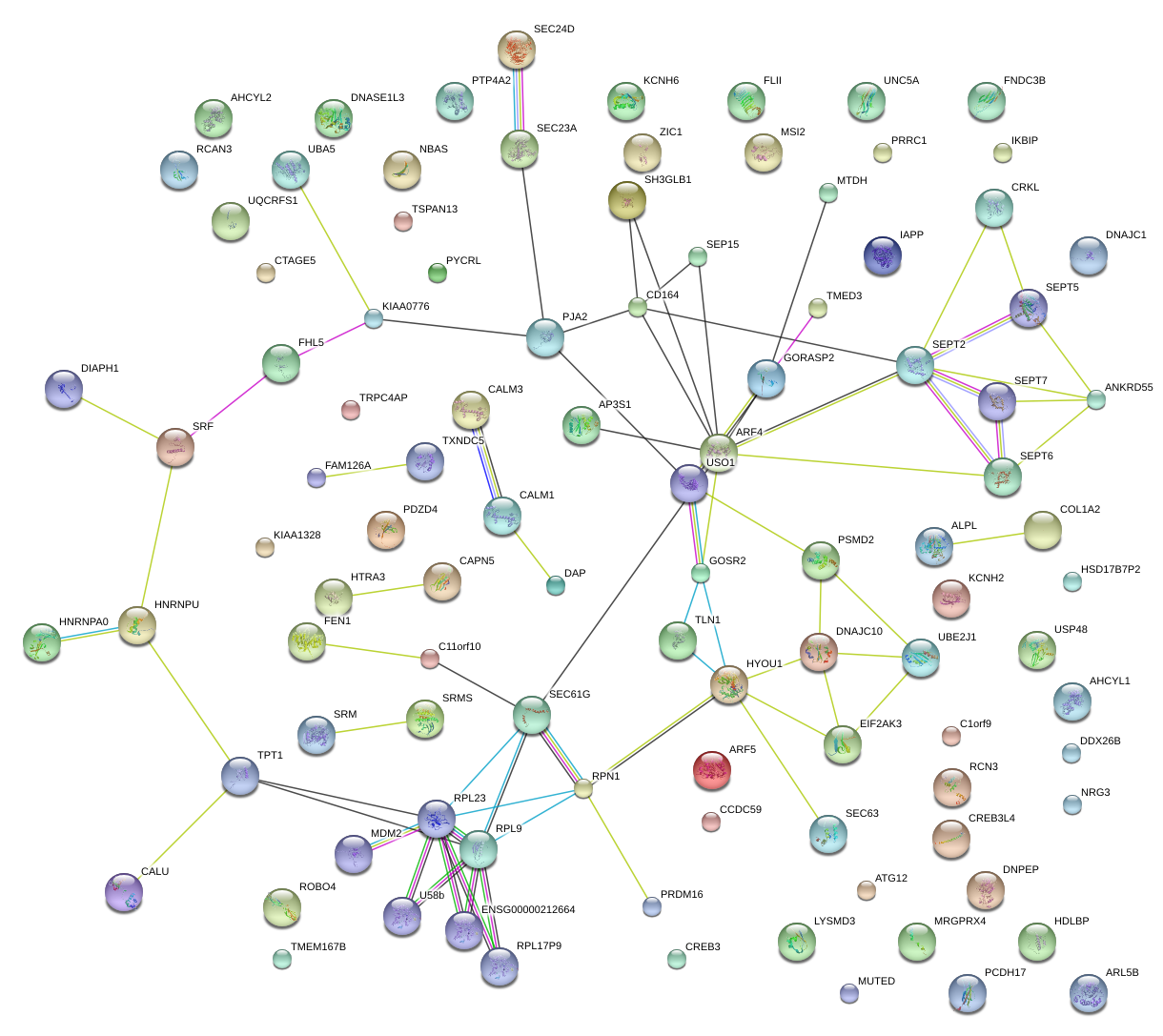
|
chr12_-_97562532
|
2.373
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chr13_+_57105859
|
1.986
|
|
PCDH17
|
protocadherin 17
|
|
chr5_+_115205515
|
1.698
|
NM_001284
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit
|
|
chr5_+_115205472
|
1.386
|
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit
|
|
chr7_+_93861808
|
1.369
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr3_+_173240983
|
1.336
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr6_-_109809595
|
1.320
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr10_+_18988282
|
1.270
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr7_+_16759845
|
1.220
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr4_+_76868763
|
1.217
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr6_-_109810246
|
1.209
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr19_+_54723044
|
1.209
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr19_+_51796399
|
1.127
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr4_+_76868830
|
1.121
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr7_+_128166581
|
1.086
|
NM_001130674
NM_001219
|
CALU
|
calumenin
|
|
chr19_+_54996613
|
1.074
|
|
|
|
|
chr2_-_15618896
|
1.068
|
NM_015909
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr6_-_108385905
|
1.050
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr7_+_128166631
|
1.009
|
|
CALU
|
calumenin
|
|
chr2_-_15618865
|
1.006
|
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr4_+_76868850
|
0.982
|
NM_003715
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr10_-_18988172
|
0.961
|
|
|
|
|
chr1_-_87152372
|
0.946
|
|
SEP15
|
15 kDa selenoprotein
|
|
chr7_-_150305933
|
0.919
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr6_-_108386000
|
0.915
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr9_-_35722226
|
0.911
|
|
TLN1
|
talin 1
|
|
chr9_+_35722207
|
0.909
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr11_+_61316725
|
0.884
|
NM_004111
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr18_-_45272840
|
0.879
|
NM_000985
NM_001035006
|
RPL17
|
ribosomal protein L17
|
|
chr10_-_22332368
|
0.875
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr18_-_45272820
|
0.869
|
|
RPL17
|
ribosomal protein L17
|
|
chr6_-_108386013
|
0.861
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr1_+_110328955
|
0.855
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_-_87152396
|
0.847
|
|
SEP15
|
15 kDa selenoprotein
|
|
chr9_-_35722175
|
0.818
|
|
TLN1
|
talin 1
|
|
chr5_-_137116746
|
0.815
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr7_+_128166636
|
0.808
|
|
CALU
|
calumenin
|
|
chr18_-_45272775
|
0.806
|
|
RPL17
|
ribosomal protein L17
|
|
chr11_-_118433061
|
0.804
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr7_-_54794358
|
0.801
|
|
SEC61G
|
Sec61 gamma subunit
|
|
chr6_-_108386071
|
0.797
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr4_-_76868581
|
0.796
|
|
|
|
|
chr6_-_108386059
|
0.794
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr2_-_88707978
|
0.790
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr1_-_32176475
|
0.777
|
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr1_-_21982192
|
0.773
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr4_-_119976631
|
0.772
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr1_-_21982037
|
0.770
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr11_-_118433043
|
0.769
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr19_+_51796330
|
0.763
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chrX_+_134482387
|
0.760
|
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr5_-_115205143
|
0.755
|
NM_004707
|
ATG12
|
ATG12 autophagy related 12 homolog (S. cerevisiae)
|
|
chr1_+_21708428
|
0.749
|
NM_000478
NM_001127501
NM_001177520
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chr7_+_128166664
|
0.745
|
|
CALU
|
calumenin
|
|
chr2_-_241903657
|
0.743
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr2_+_183289205
|
0.736
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr2_+_171494026
|
0.731
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr1_-_243094233
|
0.722
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr6_-_108386102
|
0.714
|
NM_007214
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr9_-_35722389
|
0.700
|
NM_006289
|
TLN1
|
talin 1
|
|
chr12_-_97562836
|
0.698
|
|
IKBIP
|
IKBKB interacting protein
|
|
chr10_+_83625049
|
0.690
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr11_-_118432966
|
0.687
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr11_+_61316928
|
0.686
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr1_-_11042668
|
0.682
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr4_-_39136448
|
0.680
|
NM_001024921
|
RPL9
|
ribosomal protein L9
|
|
chr9_-_35722115
|
0.676
|
|
TLN1
|
talin 1
|
|
chr4_+_8322358
|
0.672
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr3_-_129852182
|
0.666
|
|
RPN1
|
ribophorin I
|
|
chr14_-_38642029
|
0.661
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr1_-_11042663
|
0.659
|
|
SRM
|
spermidine synthase
|
|
chr5_+_126881207
|
0.656
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr5_-_10814179
|
0.656
|
|
DAP
|
death-associated protein
|
|
chr2_+_241903395
|
0.647
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr20_-_33144262
|
0.645
|
NM_015638
NM_199368
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr5_-_89861048
|
0.642
|
NM_198273
|
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3
|
|
chr11_-_118433025
|
0.637
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr22_+_19601721
|
0.621
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr11_+_61316968
|
0.618
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr6_-_7855125
|
0.618
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr3_-_57558193
|
0.612
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr7_+_128166688
|
0.609
|
|
CALU
|
calumenin
|
|
chr14_-_38642165
|
0.608
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr1_+_86943074
|
0.598
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr1_+_110329224
|
0.595
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr4_-_119976736
|
0.589
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr17_+_42355492
|
0.582
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr19_-_34395947
|
0.582
|
NM_006003
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr1_-_11042634
|
0.579
|
|
SRM
|
spermidine synthase
|
|
chr7_-_23020199
|
0.573
|
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr11_-_1975144
|
0.569
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr5_-_10814341
|
0.564
|
|
DAP
|
death-associated protein
|
|
chr15_+_77390573
|
0.563
|
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr18_+_32663056
|
0.561
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr9_-_35722089
|
0.560
|
|
TLN1
|
talin 1
|
|
chr22_+_19601619
|
0.559
|
NM_005207
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr12_+_67489063
|
0.559
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr1_-_21982256
|
0.557
|
NM_001032730
NM_032236
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr6_-_90119158
|
0.555
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr17_+_52689372
|
0.552
|
NM_170721
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr11_-_118433082
|
0.550
|
NM_006389
NM_001130991
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr6_-_90119068
|
0.547
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr3_+_148610516
|
0.540
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr1_+_110329279
|
0.536
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr20_-_33144013
|
0.529
|
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr11_-_61316628
|
0.527
|
NM_014206
|
C11orf10
|
chromosome 11 open reading frame 10
|
|
chr3_+_133861894
|
0.526
|
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr1_+_109434896
|
0.524
|
NM_020141
|
TMEM167B
|
transmembrane protein 167B
|
|
chr12_-_81276285
|
0.523
|
NM_014167
|
CCDC59
|
coiled-coil domain containing 59
|
|
chr5_-_108773468
|
0.521
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr11_+_61316989
|
0.521
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr1_-_11042649
|
0.518
|
|
SRM
|
spermidine synthase
|
|
chr15_+_77390552
|
0.507
|
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr8_+_98725604
|
0.503
|
|
MTDH
|
metadherin
|
|
chr1_-_32176275
|
0.501
|
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr13_-_44813308
|
0.498
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr5_-_137117048
|
0.497
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr1_+_170768635
|
0.496
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr13_-_44813277
|
0.496
|
NM_003295
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr6_+_97076451
|
0.495
|
|
KIAA0776
|
KIAA0776
|
|
chr5_-_140978744
|
0.493
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr11_-_1974992
|
0.492
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr14_+_38806028
|
0.490
|
NM_005930
NM_203355
|
CTAGE5
|
CTAGE family, member 5
|
|
chr1_+_170768914
|
0.487
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr3_-_178396892
|
0.486
|
|
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1
|
|
chr20_-_33144254
|
0.486
|
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr19_-_51666503
|
0.485
|
|
PNMAL1
|
PNMA-like 1
|
|
chr5_+_118434620
|
0.484
|
|
DMXL1
|
Dmx-like 1
|
|
chr15_-_53667800
|
0.484
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr4_-_83569476
|
0.483
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr1_-_87152596
|
0.478
|
NM_004261
NM_203341
|
SEP15
|
15 kDa selenoprotein
|
|
chr2_+_173129022
|
0.475
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr18_-_32662942
|
0.472
|
|
C18orf10
|
chromosome 18 open reading frame 10
|
|
chr1_+_110328922
|
0.472
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr4_+_129201933
|
0.471
|
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr18_-_32662946
|
0.467
|
|
C18orf10
|
chromosome 18 open reading frame 10
|
|
chr6_-_90119008
|
0.466
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr7_-_26206891
|
0.465
|
NM_002137
NM_031243
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr7_+_96584929
|
0.463
|
|
ACN9
|
ACN9 homolog (S. cerevisiae)
|
|
chr7_-_23020257
|
0.460
|
NM_032581
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr4_+_3737872
|
0.460
|
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr16_+_2023279
|
0.459
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr9_-_101024054
|
0.458
|
NM_033087
|
ALG2
|
asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae)
|
|
chr14_-_91576037
|
0.457
|
NM_004239
|
TRIP11
|
thyroid hormone receptor interactor 11
|
|
chr20_+_48241334
|
0.457
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr19_-_51608665
|
0.456
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr20_+_30871438
|
0.456
|
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr18_-_32663082
|
0.450
|
NM_015476
|
C18orf10
|
chromosome 18 open reading frame 10
|
|
chr7_-_30032649
|
0.448
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr1_+_26631359
|
0.446
|
NM_024887
NM_205861
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr6_-_106880182
|
0.446
|
|
ATG5
|
ATG5 autophagy related 5 homolog (S. cerevisiae)
|
|
chr6_+_43705254
|
0.440
|
NM_001003690
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chr16_-_28130201
|
0.439
|
|
XPO6
|
exportin 6
|
|
chr15_+_77390592
|
0.439
|
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr10_+_17312654
|
0.437
|
|
VIM
|
vimentin
|
|
chr2_-_69467825
|
0.431
|
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr1_+_144287344
|
0.431
|
NM_006099
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr17_+_42355482
|
0.430
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr5_-_140978747
|
0.427
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr1_+_110328941
|
0.425
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr6_+_97076390
|
0.425
|
NM_015323
|
KIAA0776
|
KIAA0776
|
|
chr1_+_110328830
|
0.425
|
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr18_-_32662843
|
0.423
|
|
C18orf10
|
chromosome 18 open reading frame 10
|
|
chr11_-_1975629
|
0.423
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_+_110328850
|
0.422
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr14_+_101500650
|
0.420
|
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr14_+_38806280
|
0.414
|
|
CTAGE5
|
CTAGE family, member 5
|
|
chr22_+_19601723
|
0.413
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr4_-_83569572
|
0.413
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr7_-_54794393
|
0.412
|
NM_001012456
NM_014302
|
SEC61G
|
Sec61 gamma subunit
|
|
chr7_-_26206845
|
0.411
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chrX_+_77246321
|
0.411
|
NM_000291
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr3_-_129694717
|
0.409
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr8_-_9451439
|
0.407
|
|
|
|
|
chr8_+_6553278
|
0.405
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr4_+_8645421
|
0.403
|
|
CPZ
|
carboxypeptidase Z
|
|
chr5_+_43638973
|
0.403
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr19_-_34395861
|
0.403
|
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr12_+_97563049
|
0.399
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr7_-_26206889
|
0.395
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr13_-_44813203
|
0.393
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr14_+_51525942
|
0.382
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr1_+_86943103
|
0.381
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr2_-_241861175
|
0.378
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr3_+_185580554
|
0.372
|
NM_003741
|
CHRD
|
chordin
|
|
chr19_-_18252440
|
0.372
|
|
|
|
|
chr3_+_151747204
|
0.367
|
NM_032025
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr19_-_18891205
|
0.362
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr9_-_94127440
|
0.362
|
|
NOL8
|
nucleolar protein 8
|
|
chr2_+_171493870
|
0.362
|
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr8_+_98725505
|
0.361
|
NM_178812
|
MTDH
|
metadherin
|
|
chr6_+_30402203
|
0.360
|
|
|
|
|
chr11_+_113776504
|
0.359
|
|
RBM7
|
RNA binding motif protein 7
|
|
chr3_-_28365284
|
0.358
|
|
AZI2
|
5-azacytidine induced 2
|
|
chr5_-_140978751
|
0.356
|
NM_001079812
NM_005219
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr13_-_52211836
|
0.355
|
|
LECT1
|
leukocyte cell derived chemotaxin 1
|
|
chr9_+_35722490
|
0.354
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr5_-_137117333
|
0.354
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr9_-_101024031
|
0.354
|
|
ALG2
|
asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae)
|
|
chrX_+_67635593
|
0.342
|
|
YIPF6
|
Yip1 domain family, member 6
|