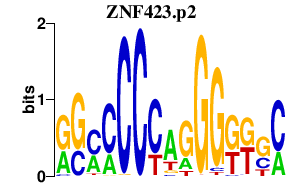
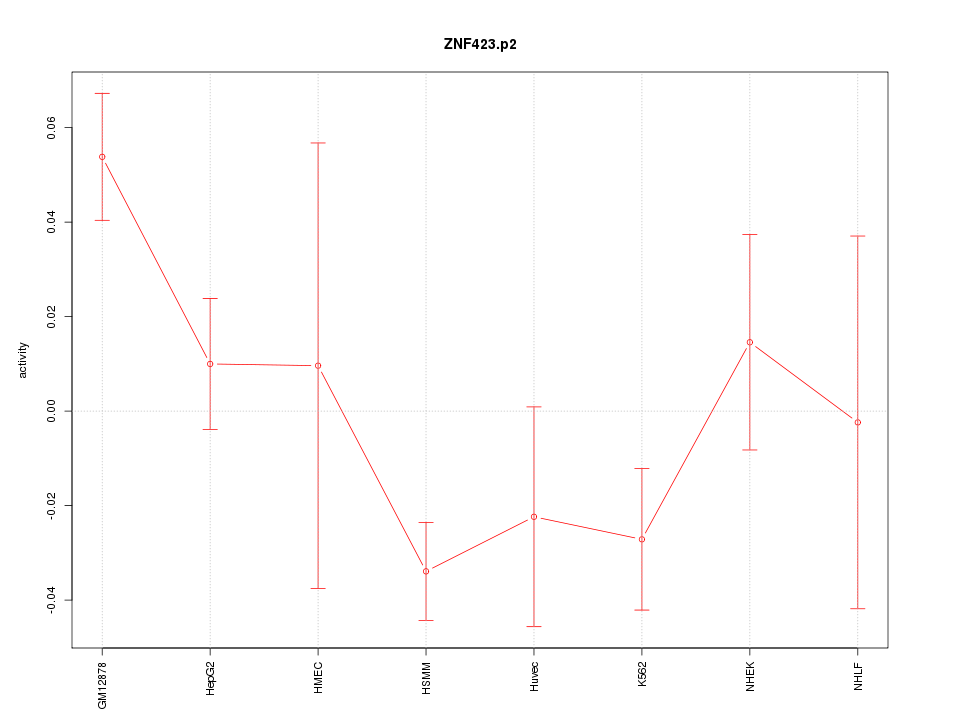
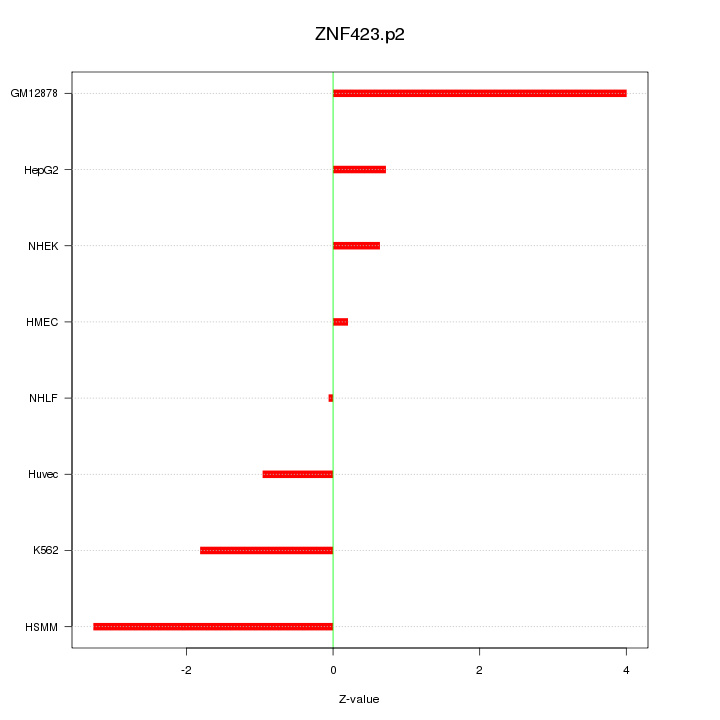
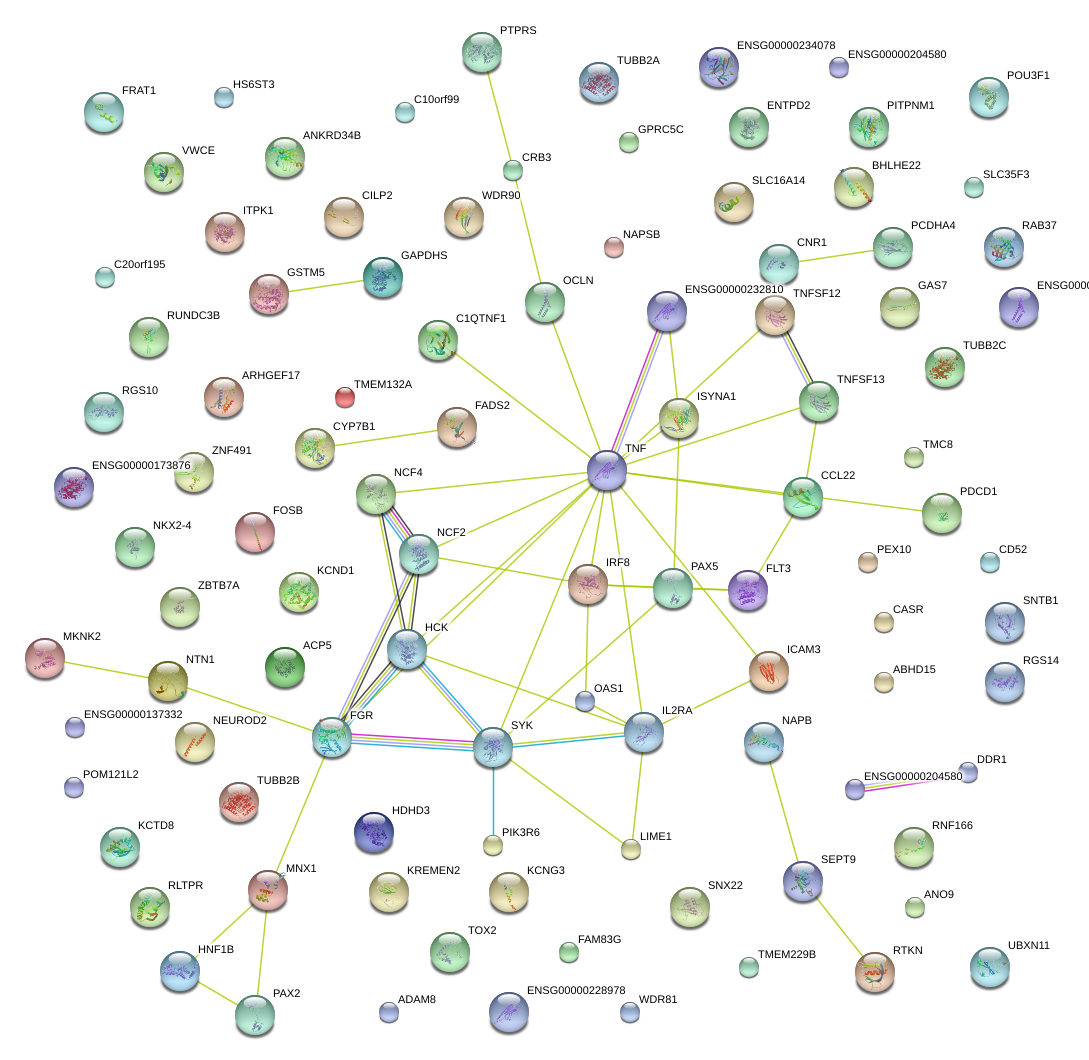
|
chr11_-_60819273
|
3.225
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr19_-_1993022
|
2.895
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr19_-_1992862
|
2.876
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr17_-_33178987
|
2.789
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr10_-_134940315
|
2.733
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr5_+_68823874
|
2.672
|
NM_002538
|
OCLN
|
occludin
|
|
chr1_-_27825178
|
2.506
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr11_-_431956
|
2.121
|
|
ANO9
|
anoctamin 9
|
|
chr10_+_85923473
|
2.029
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr19_-_11550765
|
1.962
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_-_10311262
|
1.916
|
NM_002162
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chr9_-_139068269
|
1.900
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr11_-_431995
|
1.857
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr5_+_176717449
|
1.849
|
NM_006480
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr16_+_66236512
|
1.842
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr17_-_8711701
|
1.825
|
NM_001010855
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6
|
|
chr19_-_11550576
|
1.786
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr11_-_60819110
|
1.777
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr20_-_21326046
|
1.726
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr5_+_68824143
|
1.698
|
|
OCLN
|
occludin
|
|
chr11_+_61352209
|
1.688
|
NM_004265
|
FADS2
|
fatty acid desaturase 2
|
|
chr19_+_19510056
|
1.685
|
NM_153221
|
CILP2
|
cartilage intermediate layer protein 2
|
|
chr20_+_61838479
|
1.648
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr11_+_61352348
|
1.617
|
|
FADS2
|
fatty acid desaturase 2
|
|
chr16_+_639278
|
1.601
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr17_-_33179118
|
1.583
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr8_-_65873658
|
1.541
|
NM_004820
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1
|
|
chr19_+_11770360
|
1.537
|
NM_152356
|
ZNF491
|
zinc finger protein 491
|
|
chr17_+_69950782
|
1.497
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr16_+_66236559
|
1.487
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr6_-_3172775
|
1.450
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B
|
|
chr3_+_123385870
|
1.440
|
NM_001178065
|
CASR
|
calcium-sensing receptor
|
|
chr17_+_70244945
|
1.438
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr1_-_38285033
|
1.430
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr16_+_2954217
|
1.420
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr17_+_7402788
|
1.413
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr17_-_18848713
|
1.397
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr2_-_42574587
|
1.394
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr19_+_19510098
|
1.375
|
|
CILP2
|
cartilage intermediate layer protein 2
|
|
chr20_+_61838389
|
1.363
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr1_-_181826086
|
1.361
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr11_+_60448487
|
1.333
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr19_-_4017739
|
1.328
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr13_+_95541093
|
1.303
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr19_-_18409788
|
1.296
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr6_-_88932485
|
1.287
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr4_-_44145266
|
1.285
|
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr14_-_67051773
|
1.268
|
NM_182526
|
TMEM229B
|
transmembrane protein 229B
|
|
chr1_+_110056386
|
1.263
|
NM_000851
|
GSTM5
|
glutathione S-transferase mu 5
|
|
chr10_+_99069007
|
1.256
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr17_-_35017650
|
1.250
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr9_+_92604025
|
1.231
|
NM_001174168
|
SYK
|
spleen tyrosine kinase
|
|
chr6_+_30959839
|
1.226
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr20_+_41978195
|
1.224
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr17_+_1574583
|
1.224
|
NM_001163811
|
WDR81
|
WD repeat domain 81
|
|
chr10_-_6144119
|
1.219
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr11_+_72696963
|
1.217
|
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr9_-_37024358
|
1.212
|
NM_016734
|
PAX5
|
paired box 5
|
|
chr12_+_111829191
|
1.208
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr10_-_121286034
|
1.201
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr14_-_92651896
|
1.199
|
NM_001142593
NM_001142594
NM_014216
|
ITPK1
|
inositol 1,3,4-triphosphate 5/6 kinase
|
|
chr15_+_62230962
|
1.189
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr9_+_92603920
|
1.171
|
|
SYK
|
spleen tyrosine kinase
|
|
chr2_-_120697382
|
1.167
|
|
TMEM185B
|
transmembrane protein 185B (pseudogene)
|
|
chr19_+_50663089
|
1.167
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr6_-_27387989
|
1.166
|
NM_033482
|
POM121L2
|
POM121 membrane glycoprotein-like 2
|
|
chr5_-_79901838
|
1.160
|
|
ANKRD34B
|
ankyrin repeat domain 34B
|
|
chr19_+_40716153
|
1.158
|
NM_014364
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic
|
|
chr1_+_26517032
|
1.157
|
|
CD52
|
CD52 molecule
|
|
chr19_+_6415259
|
1.156
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr19_-_18409971
|
1.154
|
NM_001170938
NM_016368
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr1_-_26517342
|
1.153
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr8_+_65656164
|
1.145
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr2_-_120697441
|
1.144
|
|
TMEM185B
|
transmembrane protein 185B (pseudogene)
|
|
chr5_+_176717553
|
1.142
|
|
|
|
|
chr17_-_10042579
|
1.126
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr2_-_120697148
|
1.120
|
|
TMEM185B
|
transmembrane protein 185B (pseudogene)
|
|
chr1_+_26517045
|
1.114
|
|
CD52
|
CD52 molecule
|
|
chr6_+_31651319
|
1.109
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr11_-_67028415
|
1.098
|
|
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1
|
|
chr2_-_74522563
|
1.092
|
NM_001015055
|
RTKN
|
rhotekin
|
|
chr17_-_35017691
|
1.092
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr17_+_70244553
|
1.082
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr17_+_74531873
|
1.081
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr16_+_55950195
|
1.073
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr11_+_72697310
|
1.072
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr17_+_8865547
|
1.063
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr2_-_74522534
|
1.058
|
|
RTKN
|
rhotekin
|
|
chr7_+_87095640
|
1.057
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chrX_-_48713194
|
1.056
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr7_-_156495985
|
1.043
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr16_-_87297525
|
1.041
|
NM_001171816
|
RNF166
|
ring finger protein 166
|
|
chr1_-_2333800
|
1.037
|
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr19_-_44214707
|
1.034
|
|
|
|
|
chr17_+_73638430
|
1.033
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr2_-_230641862
|
1.029
|
NM_152527
|
SLC16A14
|
solute carrier family 16, member 14 (monocarboxylic acid transporter 14)
|
|
chr9_+_92603884
|
1.020
|
|
SYK
|
spleen tyrosine kinase
|
|
chr20_-_23350038
|
1.006
|
NM_022080
|
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta
|
|
chr9_+_135880363
|
1.003
|
|
NCRNA00094
|
non-protein coding RNA 94
|
|
chr16_+_84490267
|
1.002
|
NM_002163
|
IRF8
|
interferon regulatory factor 8
|
|
chr20_+_61654816
|
1.002
|
NM_024059
|
C20orf195
|
chromosome 20 open reading frame 195
|
|
chr12_+_111829121
|
0.996
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr9_-_115178161
|
0.986
|
NM_031219
|
HDHD3
|
haloacid dehalogenase-like hydrolase domain containing 3
|
|
chr19_-_5291700
|
0.966
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr2_-_242449662
|
0.962
|
NM_005018
|
PDCD1
|
programmed cell death 1
|
|
chr13_-_27572646
|
0.962
|
NM_004119
|
FLT3
|
fms-related tyrosine kinase 3
|
|
chr2_-_74522513
|
0.959
|
|
RTKN
|
rhotekin
|
|
chr8_-_121892877
|
0.957
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr20_+_30103642
|
0.947
|
NM_001172129
NM_001172130
NM_001172131
NM_001172132
NM_001172133
NM_002110
|
HCK
|
hemopoietic cell kinase
|
|
chr1_+_232416618
|
0.945
|
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr17_-_24918052
|
0.944
|
NM_198147
|
ABHD15
|
abhydrolase domain containing 15
|
|
chr22_+_35586950
|
0.940
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr2_+_46623325
|
0.940
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr6_+_33352886
|
0.934
|
NM_003782
|
B3GALT4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4
|
|
chr3_+_13496714
|
0.929
|
NM_024827
NM_001136041
|
HDAC11
|
histone deacetylase 11
|
|
chr1_+_110031940
|
0.928
|
NM_000561
NM_146421
|
GSTM1
|
glutathione S-transferase mu 1
|
|
chr9_+_92603894
|
0.917
|
|
SYK
|
spleen tyrosine kinase
|
|
chr6_+_31538934
|
0.917
|
NM_006674
|
HCP5
|
HLA complex P5
|
|
chr20_+_35964962
|
0.909
|
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr16_-_11588525
|
0.902
|
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr20_-_43972982
|
0.902
|
|
PLTP
|
phospholipid transfer protein
|
|
chr7_+_26158257
|
0.900
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chrX_+_49013424
|
0.896
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr14_+_92049396
|
0.887
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr12_+_439803
|
0.885
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr14_+_23508922
|
0.884
|
NM_001193636
NM_001193637
NM_001193635
|
DHRS4L2
DHRS4L1
|
dehydrogenase/reductase (SDR family) member 4 like 2
dehydrogenase/reductase (SDR family) member 4 like 1
|
|
chr3_+_152287365
|
0.883
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr9_+_134275431
|
0.870
|
NM_207417
|
C9orf171
|
chromosome 9 open reading frame 171
|
|
chr11_-_47164514
|
0.862
|
NM_001184974
NM_016223
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr3_-_16530132
|
0.857
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr8_-_70908130
|
0.852
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr10_-_76665691
|
0.847
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr5_+_150380325
|
0.843
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr19_-_1997229
|
0.835
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr2_-_144991386
|
0.826
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_-_129024665
|
0.820
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr2_+_202607554
|
0.812
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr1_-_2333863
|
0.811
|
NM_002617
NM_153818
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr6_+_73388529
|
0.808
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr7_+_128615948
|
0.794
|
NM_005631
|
SMO
|
smoothened homolog (Drosophila)
|
|
chr7_-_29995809
|
0.792
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr19_-_44214972
|
0.791
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chr15_-_88095363
|
0.791
|
NM_018670
|
MESP1
|
mesoderm posterior 1 homolog (mouse)
|
|
chr20_+_43531806
|
0.786
|
NM_006103
|
WFDC2
|
WAP four-disulfide core domain 2
|
|
chr17_+_7402315
|
0.781
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr1_-_9052253
|
0.778
|
NM_001135585
NM_003039
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr21_-_45786709
|
0.775
|
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr8_+_145137354
|
0.774
|
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chr1_-_210939820
|
0.774
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr21_+_25856311
|
0.772
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr5_+_150380270
|
0.772
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr5_+_74197473
|
0.768
|
|
LOC441086
|
hypothetical LOC441086
|
|
chr20_-_51633042
|
0.764
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr19_-_56841819
|
0.763
|
NM_001098612
|
SIGLEC14
|
sialic acid binding Ig-like lectin 14
|
|
chr1_+_26516967
|
0.758
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr11_-_61415429
|
0.752
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr17_-_2187427
|
0.749
|
NM_018128
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr7_+_99919485
|
0.747
|
NM_173564
|
C7orf51
|
chromosome 7 open reading frame 51
|
|
chr3_+_185430910
|
0.747
|
NM_138345
|
VWA5B2
|
von Willebrand factor A domain containing 5B2
|
|
chr8_-_11361594
|
0.743
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr20_+_43895802
|
0.741
|
NM_001042632
NM_001042633
NM_033421
NM_152897
|
SNX21
|
sorting nexin family member 21
|
|
chr17_-_77488258
|
0.733
|
NM_006907
NM_153824
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr16_-_10183616
|
0.731
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr4_-_44145580
|
0.729
|
NM_198353
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr3_-_16529996
|
0.723
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr19_+_43502360
|
0.722
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr1_+_9634368
|
0.716
|
NM_005026
|
PIK3CD
|
phosphoinositide-3-kinase, catalytic, delta polypeptide
|
|
chr20_-_43972949
|
0.713
|
|
PLTP
|
phospholipid transfer protein
|
|
chr19_+_2792432
|
0.713
|
NM_001172775
NM_152791
|
ZNF555
|
zinc finger protein 555
|
|
chr19_+_43502380
|
0.712
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr14_+_103065226
|
0.711
|
NM_152307
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr5_+_150380173
|
0.710
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr17_+_45993338
|
0.709
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr19_-_1989937
|
0.708
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr19_+_17440577
|
0.707
|
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1
|
|
chr19_-_12128528
|
0.707
|
NM_145233
|
ZNF625-ZNF20
ZNF625
|
ZNF625-ZNF20 read-through transcript
zinc finger protein 625
|
|
chr7_+_27102508
|
0.704
|
|
|
|
|
chr9_+_134027154
|
0.704
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chr8_-_101227203
|
0.700
|
NM_001029860
|
FBXO43
|
F-box protein 43
|
|
chr3_-_49732062
|
0.699
|
NM_198722
|
AMIGO3
|
adhesion molecule with Ig-like domain 3
|
|
chr14_+_91859904
|
0.697
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr12_+_63504618
|
0.692
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr8_+_22479115
|
0.691
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr14_+_105024576
|
0.685
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr11_-_75057062
|
0.685
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chr9_-_132804243
|
0.684
|
NM_001145106
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr17_+_74531820
|
0.683
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr14_+_103065288
|
0.679
|
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr12_+_63504708
|
0.679
|
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr8_+_121892719
|
0.676
|
|
|
|
|
chr22_-_49315003
|
0.672
|
NM_001113756
|
TYMP
|
thymidine phosphorylase
|
|
chr11_-_67153959
|
0.670
|
NM_181843
|
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8
|
|
chr1_-_226530350
|
0.668
|
|
|
|
|
chr18_+_75261231
|
0.664
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr15_+_22751162
|
0.661
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr19_+_7647934
|
0.658
|
NM_174918
|
C19orf59
|
chromosome 19 open reading frame 59
|
|
chr11_-_66877519
|
0.652
|
NM_021173
|
POLD4
|
polymerase (DNA-directed), delta 4
|
|
chr19_-_13978096
|
0.651
|
NM_002918
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr3_+_198214173
|
0.649
|
|
LOC100507057
MFI2
|
hypothetical LOC100507057
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5
|
|
chr16_+_31027249
|
0.647
|
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|