Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for MAFF_MAFG
Z-value: 1.66
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.7 | MAFF |
|
MAFG
|
ENSG00000197063.6 | MAFG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFF | hg19_v2_chr22_+_38609538_38609547, hg19_v2_chr22_+_38597889_38598021 | 0.48 | 5.7e-02 | Click! |
| MAFG | hg19_v2_chr17_-_79881408_79881423, hg19_v2_chr17_-_79885576_79885624 | 0.26 | 3.4e-01 | Click! |
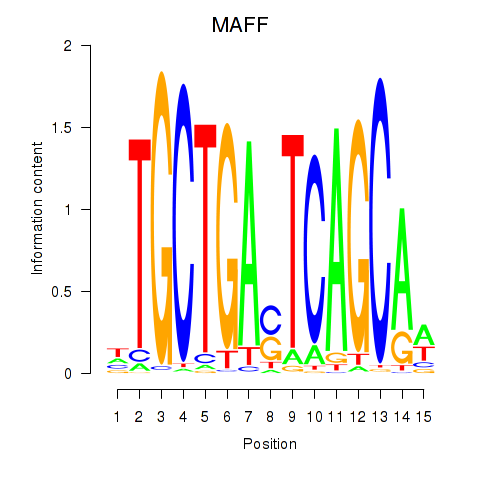
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709527 | 19.89 |
ENST00000250784.8 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chrY_+_2709906 | 7.15 |
ENST00000430575.1 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr3_-_99594948 | 4.83 |
ENST00000471562.1 ENST00000495625.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr3_-_99595037 | 4.51 |
ENST00000383694.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr22_+_21133469 | 4.40 |
ENST00000406799.1 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr16_+_30211181 | 2.62 |
ENST00000395138.2 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr1_-_110283138 | 2.55 |
ENST00000256594.3 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr5_-_42811986 | 2.23 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr4_+_74275057 | 2.22 |
ENST00000511370.1 |
ALB |
albumin |
| chr10_-_90751038 | 2.18 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr17_-_76921459 | 2.14 |
ENST00000262768.7 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr5_-_42812143 | 2.13 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr15_+_58724184 | 2.09 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr17_-_63822563 | 1.97 |
ENST00000317442.8 |
CEP112 |
centrosomal protein 112kDa |
| chr5_-_111093167 | 1.81 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr7_+_100199800 | 1.62 |
ENST00000223061.5 |
PCOLCE |
procollagen C-endopeptidase enhancer |
| chr12_-_91573132 | 1.58 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr5_-_111093406 | 1.47 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr16_+_56685796 | 1.46 |
ENST00000334346.2 ENST00000562399.1 |
MT1B |
metallothionein 1B |
| chr5_-_111092873 | 1.45 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr17_+_53343577 | 1.43 |
ENST00000573945.1 |
HLF |
hepatic leukemia factor |
| chr17_-_76899275 | 1.43 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr15_+_33010175 | 1.36 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr20_+_61287711 | 1.35 |
ENST00000370507.1 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
| chr2_-_169104651 | 1.31 |
ENST00000355999.4 |
STK39 |
serine threonine kinase 39 |
| chr17_-_10325261 | 1.25 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr7_+_89975979 | 1.25 |
ENST00000257659.8 ENST00000222511.6 ENST00000417207.1 |
GTPBP10 |
GTP-binding protein 10 (putative) |
| chr2_-_175629135 | 1.22 |
ENST00000409542.1 ENST00000409219.1 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr16_+_82068830 | 1.19 |
ENST00000199936.4 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr7_-_22234381 | 1.15 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_-_190044480 | 1.14 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr2_+_102953608 | 1.11 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr8_+_26240414 | 1.07 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr5_-_111093081 | 1.05 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr1_+_196788887 | 1.01 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr2_-_11484710 | 0.96 |
ENST00000315872.6 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
| chr10_+_54074033 | 0.92 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr10_+_5005445 | 0.92 |
ENST00000380872.4 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr12_+_10365404 | 0.91 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr16_-_25122785 | 0.90 |
ENST00000563962.1 ENST00000569920.1 |
RP11-449H11.1 |
RP11-449H11.1 |
| chr1_-_203155868 | 0.89 |
ENST00000255409.3 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr5_-_111093340 | 0.89 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr16_+_14802801 | 0.88 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr16_-_87970122 | 0.87 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr17_+_27369918 | 0.86 |
ENST00000323372.4 |
PIPOX |
pipecolic acid oxidase |
| chr16_-_24942273 | 0.85 |
ENST00000571406.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr7_-_122526799 | 0.85 |
ENST00000334010.7 ENST00000313070.7 |
CADPS2 |
Ca++-dependent secretion activator 2 |
| chr5_-_149535421 | 0.84 |
ENST00000261799.4 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
| chr4_+_3768075 | 0.82 |
ENST00000509482.1 ENST00000330055.5 |
ADRA2C |
adrenoceptor alpha 2C |
| chr12_+_111051902 | 0.81 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr16_-_28608364 | 0.81 |
ENST00000533150.1 |
SULT1A2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr17_+_60758814 | 0.81 |
ENST00000579432.1 ENST00000446119.2 |
MRC2 |
mannose receptor, C type 2 |
| chr19_+_535835 | 0.80 |
ENST00000607527.1 ENST00000606065.1 |
CDC34 |
cell division cycle 34 |
| chr4_+_26585538 | 0.79 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr17_-_64187973 | 0.79 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr6_-_90529418 | 0.79 |
ENST00000439638.1 ENST00000369393.3 ENST00000428876.1 |
MDN1 |
MDN1, midasin homolog (yeast) |
| chr22_+_44319648 | 0.78 |
ENST00000423180.2 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr1_+_17248418 | 0.77 |
ENST00000375541.5 |
CROCC |
ciliary rootlet coiled-coil, rootletin |
| chr5_+_35856951 | 0.76 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr4_-_170924888 | 0.74 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr19_-_23578220 | 0.72 |
ENST00000595533.1 ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91 |
zinc finger protein 91 |
| chr2_-_211168332 | 0.72 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr6_-_47277634 | 0.70 |
ENST00000296861.2 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
| chr15_-_72668185 | 0.68 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr6_+_131894284 | 0.68 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr19_-_36297348 | 0.66 |
ENST00000589835.1 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr3_-_125802765 | 0.65 |
ENST00000514891.1 ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr19_+_16254488 | 0.64 |
ENST00000588246.1 ENST00000593031.1 |
HSH2D |
hematopoietic SH2 domain containing |
| chr17_+_79650962 | 0.63 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr17_+_48243352 | 0.63 |
ENST00000344627.6 ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA |
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr6_-_109330702 | 0.60 |
ENST00000356644.7 |
SESN1 |
sestrin 1 |
| chr17_+_66508154 | 0.59 |
ENST00000358598.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr3_-_178969403 | 0.58 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr17_+_66509019 | 0.57 |
ENST00000585981.1 ENST00000589480.1 ENST00000585815.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr17_-_28618948 | 0.57 |
ENST00000261714.6 |
BLMH |
bleomycin hydrolase |
| chr2_-_167232484 | 0.55 |
ENST00000375387.4 ENST00000303354.6 ENST00000409672.1 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
| chr17_+_66508537 | 0.55 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr2_+_211421262 | 0.55 |
ENST00000233072.5 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chrX_-_10544942 | 0.54 |
ENST00000380779.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr19_-_43383789 | 0.54 |
ENST00000595356.1 |
PSG1 |
pregnancy specific beta-1-glycoprotein 1 |
| chr1_+_183774240 | 0.53 |
ENST00000360851.3 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr21_-_27543425 | 0.53 |
ENST00000448388.2 |
APP |
amyloid beta (A4) precursor protein |
| chr16_+_56716336 | 0.52 |
ENST00000394485.4 ENST00000562939.1 |
MT1X |
metallothionein 1X |
| chr16_-_56701933 | 0.51 |
ENST00000568675.1 ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G |
metallothionein 1G |
| chr14_-_51411194 | 0.51 |
ENST00000544180.2 |
PYGL |
phosphorylase, glycogen, liver |
| chr3_+_45071622 | 0.51 |
ENST00000428034.1 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr4_-_52904425 | 0.47 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr12_+_51318513 | 0.47 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr10_+_124320156 | 0.47 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr6_-_25930904 | 0.46 |
ENST00000377850.3 |
SLC17A2 |
solute carrier family 17, member 2 |
| chr2_+_46844290 | 0.46 |
ENST00000238892.3 |
CRIPT |
cysteine-rich PDZ-binding protein |
| chr6_-_152489484 | 0.45 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr5_-_115152651 | 0.45 |
ENST00000250535.4 |
CDO1 |
cysteine dioxygenase type 1 |
| chr10_+_73975742 | 0.45 |
ENST00000299381.4 |
ANAPC16 |
anaphase promoting complex subunit 16 |
| chr8_+_64081118 | 0.44 |
ENST00000539294.1 |
YTHDF3 |
YTH domain family, member 3 |
| chr17_-_80231557 | 0.44 |
ENST00000392334.2 ENST00000314028.6 |
CSNK1D |
casein kinase 1, delta |
| chr19_-_45826125 | 0.43 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr4_-_16077741 | 0.43 |
ENST00000447510.2 ENST00000540805.1 ENST00000539194.1 |
PROM1 |
prominin 1 |
| chr6_-_159420780 | 0.43 |
ENST00000449822.1 |
RSPH3 |
radial spoke 3 homolog (Chlamydomonas) |
| chr11_+_34938119 | 0.43 |
ENST00000227868.4 ENST00000430469.2 ENST00000533262.1 |
PDHX |
pyruvate dehydrogenase complex, component X |
| chr9_+_134378289 | 0.42 |
ENST00000423007.1 ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1 |
protein-O-mannosyltransferase 1 |
| chr17_-_79881408 | 0.42 |
ENST00000392366.3 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr19_-_43702231 | 0.42 |
ENST00000597374.1 ENST00000599371.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr7_-_6523755 | 0.42 |
ENST00000436575.1 ENST00000258739.4 |
DAGLB KDELR2 |
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr16_-_1429627 | 0.41 |
ENST00000248104.7 |
UNKL |
unkempt family zinc finger-like |
| chr12_+_64798095 | 0.40 |
ENST00000332707.5 |
XPOT |
exportin, tRNA |
| chrX_-_64196351 | 0.40 |
ENST00000374839.3 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chrX_-_134156502 | 0.40 |
ENST00000391440.1 |
FAM127C |
family with sequence similarity 127, member C |
| chr2_+_201981527 | 0.40 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr11_+_66276550 | 0.40 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chr1_+_196912902 | 0.39 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr18_+_11857439 | 0.39 |
ENST00000602628.1 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr12_-_71533055 | 0.39 |
ENST00000552128.1 |
TSPAN8 |
tetraspanin 8 |
| chr21_-_27423339 | 0.38 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chrX_-_64196376 | 0.37 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr14_-_23504087 | 0.37 |
ENST00000493471.2 ENST00000460922.2 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
| chrX_+_102883887 | 0.36 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr17_+_66521936 | 0.36 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_+_26585686 | 0.36 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr4_-_89080003 | 0.35 |
ENST00000237612.3 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr14_-_23504337 | 0.34 |
ENST00000361611.6 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr14_-_51411146 | 0.34 |
ENST00000532462.1 |
PYGL |
phosphorylase, glycogen, liver |
| chr9_+_131549610 | 0.34 |
ENST00000223865.8 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr2_-_166930131 | 0.34 |
ENST00000303395.4 ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A |
sodium channel, voltage-gated, type I, alpha subunit |
| chrX_-_119709637 | 0.33 |
ENST00000404115.3 |
CUL4B |
cullin 4B |
| chr10_+_88428206 | 0.33 |
ENST00000429277.2 ENST00000458213.2 ENST00000352360.5 |
LDB3 |
LIM domain binding 3 |
| chrX_-_64196307 | 0.33 |
ENST00000545618.1 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr14_-_23504432 | 0.33 |
ENST00000425762.2 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr19_+_48876300 | 0.32 |
ENST00000600863.1 ENST00000601610.1 ENST00000595322.1 |
SYNGR4 |
synaptogyrin 4 |
| chr11_-_117695449 | 0.32 |
ENST00000292079.2 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr17_-_34313685 | 0.32 |
ENST00000435911.2 ENST00000586216.1 ENST00000394509.4 |
CCL14 |
chemokine (C-C motif) ligand 14 |
| chr10_+_51187938 | 0.32 |
ENST00000311663.5 |
FAM21D |
family with sequence similarity 21, member D |
| chr9_-_130639997 | 0.32 |
ENST00000373176.1 |
AK1 |
adenylate kinase 1 |
| chr12_-_10251603 | 0.32 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr3_+_108308513 | 0.31 |
ENST00000361582.3 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr10_-_116444371 | 0.31 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr2_+_201981119 | 0.31 |
ENST00000395148.2 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr16_+_25123041 | 0.30 |
ENST00000399069.3 ENST00000380966.4 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
| chr1_+_110210644 | 0.29 |
ENST00000369831.2 ENST00000442650.1 ENST00000369827.3 ENST00000460717.3 ENST00000241337.4 ENST00000467579.3 ENST00000414179.2 ENST00000369829.2 |
GSTM2 |
glutathione S-transferase mu 2 (muscle) |
| chr20_+_61867235 | 0.29 |
ENST00000342412.6 ENST00000217169.3 |
BIRC7 |
baculoviral IAP repeat containing 7 |
| chr22_-_36635684 | 0.29 |
ENST00000358502.5 |
APOL2 |
apolipoprotein L, 2 |
| chr2_+_163200598 | 0.29 |
ENST00000437150.2 ENST00000453113.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr12_-_47219733 | 0.29 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr3_+_119499331 | 0.29 |
ENST00000393716.2 ENST00000466380.1 |
NR1I2 |
nuclear receptor subfamily 1, group I, member 2 |
| chr4_+_668348 | 0.28 |
ENST00000511290.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr17_-_28618867 | 0.28 |
ENST00000394819.3 ENST00000577623.1 |
BLMH |
bleomycin hydrolase |
| chr8_-_142011291 | 0.28 |
ENST00000521059.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr10_-_124713842 | 0.28 |
ENST00000481909.1 |
C10orf88 |
chromosome 10 open reading frame 88 |
| chr5_+_140019079 | 0.28 |
ENST00000252100.6 |
TMCO6 |
transmembrane and coiled-coil domains 6 |
| chr7_-_100844193 | 0.28 |
ENST00000440203.2 ENST00000379423.3 ENST00000223114.4 |
MOGAT3 |
monoacylglycerol O-acyltransferase 3 |
| chr4_+_667686 | 0.28 |
ENST00000505477.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr19_+_15751689 | 0.28 |
ENST00000586182.2 ENST00000591058.1 ENST00000221307.8 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr18_-_52989525 | 0.27 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr18_-_53069419 | 0.27 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr11_-_77791156 | 0.27 |
ENST00000281031.4 |
NDUFC2 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr1_-_160313025 | 0.26 |
ENST00000368069.3 ENST00000241704.7 |
COPA |
coatomer protein complex, subunit alpha |
| chr16_+_57496299 | 0.26 |
ENST00000219252.5 |
POLR2C |
polymerase (RNA) II (DNA directed) polypeptide C, 33kDa |
| chr9_-_26892765 | 0.26 |
ENST00000520187.1 ENST00000333916.5 |
CAAP1 |
caspase activity and apoptosis inhibitor 1 |
| chrX_-_107019181 | 0.26 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr7_+_139025875 | 0.26 |
ENST00000297534.6 |
C7orf55 |
chromosome 7 open reading frame 55 |
| chr12_+_105501487 | 0.26 |
ENST00000332180.5 |
KIAA1033 |
KIAA1033 |
| chr16_-_24942655 | 0.26 |
ENST00000573765.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chrX_+_129473859 | 0.25 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_+_222817629 | 0.25 |
ENST00000340535.7 |
MIA3 |
melanoma inhibitory activity family, member 3 |
| chr2_+_27651478 | 0.25 |
ENST00000379852.3 |
NRBP1 |
nuclear receptor binding protein 1 |
| chr2_+_163200848 | 0.25 |
ENST00000233612.4 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr14_+_58894141 | 0.24 |
ENST00000423743.3 |
KIAA0586 |
KIAA0586 |
| chr19_-_43383819 | 0.24 |
ENST00000312439.6 ENST00000403380.3 |
PSG1 |
pregnancy specific beta-1-glycoprotein 1 |
| chr16_+_446713 | 0.24 |
ENST00000397722.1 ENST00000454619.1 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr14_+_58894103 | 0.24 |
ENST00000354386.6 ENST00000556134.1 |
KIAA0586 |
KIAA0586 |
| chr2_-_46844242 | 0.23 |
ENST00000281382.6 |
PIGF |
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr3_-_58563094 | 0.22 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr22_-_27014043 | 0.22 |
ENST00000215939.2 |
CRYBB1 |
crystallin, beta B1 |
| chr16_+_78133536 | 0.22 |
ENST00000402655.2 ENST00000406884.2 ENST00000539474.2 ENST00000569818.1 ENST00000355860.3 ENST00000408984.3 |
WWOX |
WW domain containing oxidoreductase |
| chr3_-_113918254 | 0.22 |
ENST00000460779.1 |
DRD3 |
dopamine receptor D3 |
| chr13_-_25086879 | 0.22 |
ENST00000381989.3 |
PARP4 |
poly (ADP-ribose) polymerase family, member 4 |
| chr1_-_115212640 | 0.22 |
ENST00000393274.1 |
DENND2C |
DENN/MADD domain containing 2C |
| chr3_-_69101413 | 0.21 |
ENST00000398559.2 |
TMF1 |
TATA element modulatory factor 1 |
| chr2_-_46844159 | 0.21 |
ENST00000474980.1 ENST00000306465.4 |
PIGF |
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr4_-_165305086 | 0.21 |
ENST00000507270.1 ENST00000514618.1 ENST00000503008.1 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr20_+_3869423 | 0.21 |
ENST00000497424.1 |
PANK2 |
pantothenate kinase 2 |
| chr18_-_52989217 | 0.21 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr15_+_23810853 | 0.21 |
ENST00000568252.1 |
MKRN3 |
makorin ring finger protein 3 |
| chr2_+_70314579 | 0.21 |
ENST00000303577.5 |
PCBP1 |
poly(rC) binding protein 1 |
| chr18_-_5540471 | 0.20 |
ENST00000581833.1 ENST00000544123.1 ENST00000342933.3 ENST00000400111.3 ENST00000585142.1 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr5_+_175511859 | 0.20 |
ENST00000503724.2 ENST00000253490.4 |
FAM153B |
family with sequence similarity 153, member B |
| chr1_+_145301735 | 0.20 |
ENST00000605176.1 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr8_-_98290087 | 0.20 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr1_+_154966058 | 0.20 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr19_+_33571786 | 0.20 |
ENST00000170564.2 |
GPATCH1 |
G patch domain containing 1 |
| chr6_+_25963020 | 0.19 |
ENST00000357085.3 |
TRIM38 |
tripartite motif containing 38 |
| chr2_+_69201705 | 0.19 |
ENST00000377938.2 |
GKN1 |
gastrokine 1 |
| chr12_+_108956355 | 0.19 |
ENST00000535729.1 ENST00000431221.2 ENST00000547005.1 ENST00000311893.9 ENST00000392807.4 ENST00000338291.4 ENST00000539593.1 |
ISCU |
iron-sulfur cluster assembly enzyme |
| chr16_-_87466740 | 0.19 |
ENST00000561928.1 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
| chr2_+_90211643 | 0.19 |
ENST00000390277.2 |
IGKV3D-11 |
immunoglobulin kappa variable 3D-11 |
| chr17_+_7452189 | 0.18 |
ENST00000293825.6 |
TNFSF12 |
tumor necrosis factor (ligand) superfamily, member 12 |
| chr5_+_94890778 | 0.18 |
ENST00000380009.4 |
ARSK |
arylsulfatase family, member K |
| chr4_+_154125565 | 0.18 |
ENST00000338700.5 |
TRIM2 |
tripartite motif containing 2 |
| chr11_-_62380199 | 0.18 |
ENST00000419857.1 ENST00000394773.2 |
EML3 |
echinoderm microtubule associated protein like 3 |
| chr3_+_9404526 | 0.17 |
ENST00000452837.2 ENST00000417036.1 ENST00000419437.1 ENST00000345094.3 ENST00000515662.2 |
THUMPD3 |
THUMP domain containing 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 4.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 2.6 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.4 | 1.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 26.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.3 | 0.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 0.8 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.3 | 0.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 0.9 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.2 | 1.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 0.5 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 3.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 0.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 2.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.4 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 2.1 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 4.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 1.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.9 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 26.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 3.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 3.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 | 4.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.5 | 2.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.5 | 2.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.5 | 1.4 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 1.3 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.4 | 2.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.4 | 0.8 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.4 | 2.6 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 2.2 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.3 | 0.9 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 1.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.7 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.2 | 0.4 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.2 | 0.8 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 0.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.8 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 1.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.9 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 26.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.9 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 2.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.7 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 | 0.9 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 2.1 | GO:2000480 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of activated T cell proliferation(GO:0046007) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.4 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 1.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.8 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.3 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 0.4 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 2.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 2.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.8 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 1.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.7 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.3 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 0.2 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0098736 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.9 | GO:1903541 | regulation of exosomal secretion(GO:1903541) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.2 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 1.1 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.3 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 1.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:2000048 | positive regulation of fibrinolysis(GO:0051919) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 26.2 | GO:0005844 | polysome(GO:0005844) |
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 2.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.6 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 2.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 4.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 7.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.4 | GO:0043234 | protein complex(GO:0043234) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |