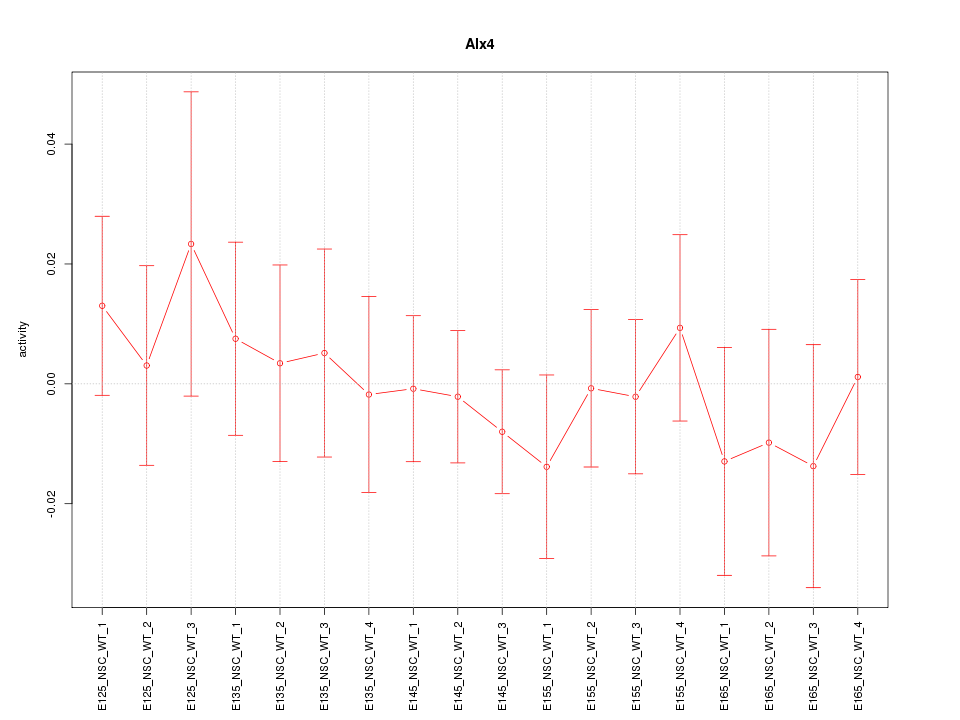
Motif ID: Alx4
Z-value: 0.530

Transcription factors associated with Alx4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Alx4 | ENSMUSG00000040310.6 | Alx4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx4 | mm10_v2_chr2_+_93642307_93642388 | -0.46 | 5.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.3 | 1.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 0.8 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.3 | 1.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.7 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 0.7 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 1.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.8 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.2 | 0.5 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.3 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 0.7 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.1 | 2.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.4 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.8 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.8 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.9 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.4 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.3 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) olefin metabolic process(GO:1900673) |
| 0.1 | 0.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.9 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 1.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.4 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.6 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.3 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.6 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.3 | GO:0015838 | proline transport(GO:0015824) amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.1 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 1.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.7 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.5 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 1.8 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.4 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.2 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.9 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.2 | GO:0051900 | regulation of mitochondrial depolarization(GO:0051900) |
| 0.0 | 0.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.4 | 1.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 0.9 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 0.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.7 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.7 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 1.6 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.3 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 2.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.1 | GO:1990393 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 0.8 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 0.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 1.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.2 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 0.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 2.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.2 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.4 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 2.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.2 | GO:0052740 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.7 | GO:0031957 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.8 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.0 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.0 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 1.5 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID_P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.4 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.9 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.7 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.4 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.5 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.7 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.8 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.8 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.9 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.0 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 2.0 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.1 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME_POL_SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.5 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.6 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |