Motif ID: Arntl_Tfe3_Mlx_Mitf_Mlxipl_Tfec
Z-value: 1.027
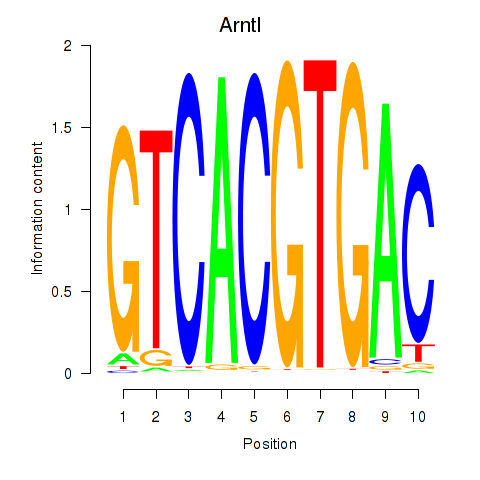
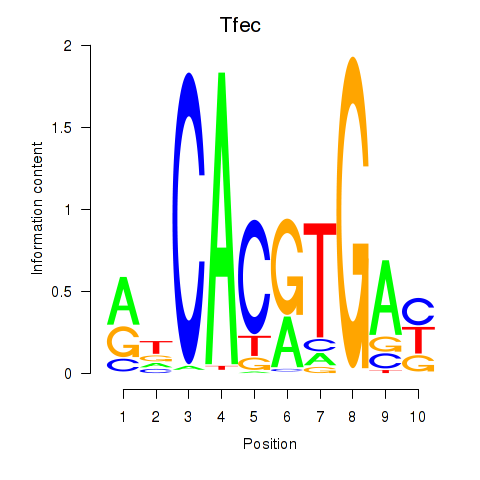
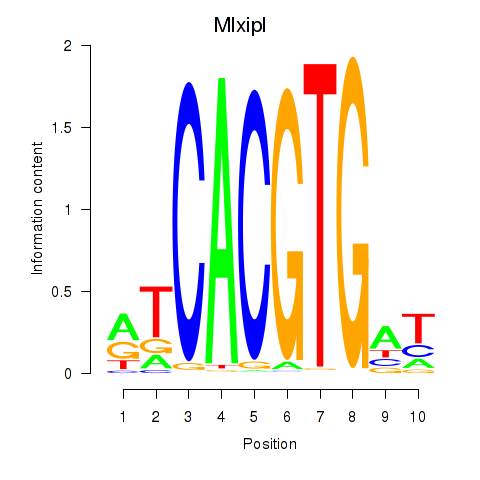
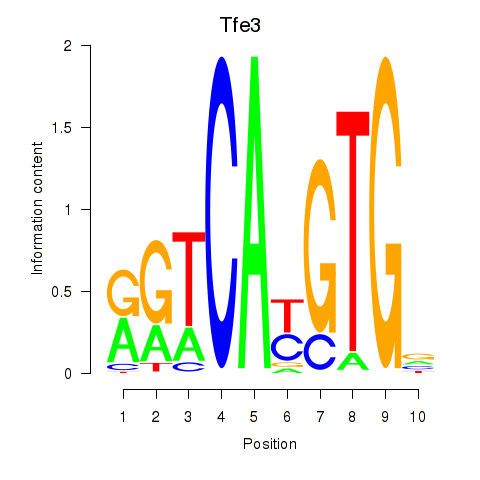
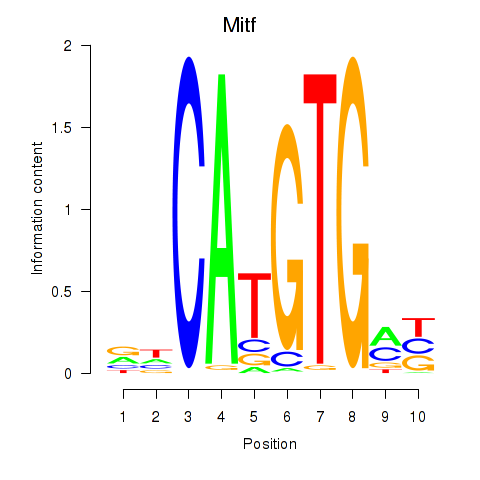
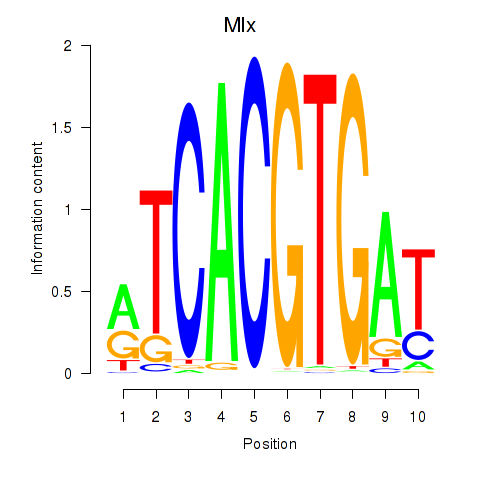
Transcription factors associated with Arntl_Tfe3_Mlx_Mitf_Mlxipl_Tfec:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Arntl | ENSMUSG00000055116.7 | Arntl |
| Mitf | ENSMUSG00000035158.9 | Mitf |
| Mlx | ENSMUSG00000017801.9 | Mlx |
| Mlxipl | ENSMUSG00000005373.7 | Mlxipl |
| Tfe3 | ENSMUSG00000000134.11 | Tfe3 |
| Tfec | ENSMUSG00000029553.7 | Tfec |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mlx | mm10_v2_chr11_+_101087277_101087323 | -0.73 | 6.4e-04 | Click! |
| Arntl | mm10_v2_chr7_+_113207465_113207595 | -0.58 | 1.3e-02 | Click! |
| Tfe3 | mm10_v2_chrX_+_7762652_7762695 | -0.37 | 1.3e-01 | Click! |
| Mlxipl | mm10_v2_chr5_+_135106881_135106918 | -0.07 | 7.9e-01 | Click! |
| Mitf | mm10_v2_chr6_+_97807014_97807067 | -0.00 | 9.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0010288 | response to lead ion(GO:0010288) |
| 1.0 | 3.0 | GO:0010248 | B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.9 | 2.8 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.9 | 2.7 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.8 | 2.4 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.8 | 3.9 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.7 | 4.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.7 | 2.0 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.6 | 1.9 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.6 | 1.8 | GO:0030421 | defecation(GO:0030421) |
| 0.6 | 3.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.5 | 0.5 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.5 | 1.6 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.5 | 2.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.5 | 1.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.5 | 2.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 1.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.5 | 1.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.5 | 6.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.5 | 0.9 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.4 | 2.6 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.4 | 1.7 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 3.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 1.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.4 | 1.2 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.4 | 1.9 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.4 | 1.2 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 | 3.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.4 | 1.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.4 | 1.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.3 | 1.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 1.4 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of arachidonic acid secretion(GO:0090238) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 1.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.3 | 1.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.3 | 2.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 2.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.3 | 1.0 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.3 | 1.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.3 | 6.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.3 | 0.9 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.3 | 0.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 | 2.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.3 | 1.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 0.8 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.3 | 1.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.3 | 6.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 1.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.7 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 0.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 1.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 0.7 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 1.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.7 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 1.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 2.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.2 | 1.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.2 | 1.1 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.2 | 0.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 0.7 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 0.8 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.2 | 0.8 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of heart induction(GO:0090381) |
| 0.2 | 1.0 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.2 | 1.0 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) positive regulation of NAD(P)H oxidase activity(GO:0033864) positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.2 | 0.8 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 0.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.2 | 1.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 0.5 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 0.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 0.7 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 1.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 1.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.5 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.2 | 0.8 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.2 | 2.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.8 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 0.8 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 0.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 1.0 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 0.5 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.2 | 0.6 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.2 | 0.8 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 1.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 3.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.2 | 4.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.2 | 0.6 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 1.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.4 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.4 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.7 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 1.3 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 1.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.6 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.7 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.5 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) |
| 0.1 | 1.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 3.0 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.9 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.4 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 0.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 1.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.6 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.5 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 0.7 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.2 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.1 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.1 | 0.5 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 1.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.9 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.1 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) |
| 0.1 | 0.3 | GO:0030324 | lung development(GO:0030324) |
| 0.1 | 1.2 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.1 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.1 | 0.3 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 4.1 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.1 | 0.3 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.1 | GO:2000643 | positive regulation of vacuolar transport(GO:1903337) positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 2.4 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 0.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 1.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.6 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 1.4 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 0.1 | GO:2000319 | negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.3 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.8 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.1 | 0.3 | GO:0009204 | deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.1 | 0.5 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.4 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.1 | 1.0 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.2 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 1.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.4 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 | 1.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.4 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) negative regulation of metalloendopeptidase activity(GO:1904684) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 1.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 1.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.6 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.5 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.2 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.3 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.8 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 1.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.5 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 0.4 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.7 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.7 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.2 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 0.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.7 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.8 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.1 | GO:0002339 | B cell selection(GO:0002339) |
| 0.1 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.1 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.9 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.1 | 0.1 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.1 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 2.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 2.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.1 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.1 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 0.2 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 0.3 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.2 | GO:0048864 | stem cell development(GO:0048864) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.1 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.1 | GO:0010455 | positive regulation of cell fate commitment(GO:0010455) |
| 0.1 | 0.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.4 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 0.6 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.1 | 1.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 2.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0007521 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) muscle cell fate determination(GO:0007521) |
| 0.0 | 0.2 | GO:1990379 | lipid transport across blood brain barrier(GO:1990379) |
| 0.0 | 0.4 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.2 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.7 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.5 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.6 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.5 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.1 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 1.1 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 1.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.3 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.3 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 1.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0072610 | interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.2 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.0 | 0.2 | GO:0015786 | UDP-glucose transport(GO:0015786) protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) icosanoid biosynthetic process(GO:0046456) prostanoid biosynthetic process(GO:0046457) fatty acid derivative biosynthetic process(GO:1901570) |
| 0.0 | 2.7 | GO:1904591 | positive regulation of protein import(GO:1904591) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 2.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.0 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.4 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 1.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.0 | 0.9 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.4 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 1.3 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.5 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 1.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:1902750 | negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.3 | GO:0002385 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 0.8 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:2000191 | regulation of fatty acid transport(GO:2000191) |
| 0.0 | 0.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.4 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.3 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0019585 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:1990173 | regulation of establishment of protein localization to chromosome(GO:0070202) regulation of establishment of protein localization to telomere(GO:0070203) protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 1.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.8 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.6 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:1903671 | negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:1904738 | vascular associated smooth muscle cell migration(GO:1904738) regulation of vascular associated smooth muscle cell migration(GO:1904752) positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.1 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.0 | 0.0 | GO:0010726 | regulation of hydrogen peroxide metabolic process(GO:0010310) positive regulation of hydrogen peroxide metabolic process(GO:0010726) regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0090169 | regulation of spindle assembly(GO:0090169) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0034244 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.1 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.5 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0060768 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.0 | 0.0 | GO:0036124 | histone H3-K9 trimethylation(GO:0036124) |
| 0.0 | 0.4 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.0 | GO:0086103 | G-protein coupled receptor signaling pathway involved in heart process(GO:0086103) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of endodermal cell differentiation(GO:1903225) negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.9 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.7 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.0 | 2.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.9 | 6.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.8 | 4.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.7 | 3.0 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.5 | 1.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.5 | 1.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 2.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 2.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 2.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 5.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 1.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.3 | 0.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 3.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 1.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 1.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 1.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 0.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 0.6 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 3.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.2 | 0.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 0.7 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 1.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 0.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 1.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 1.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 2.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 1.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 3.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 0.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 1.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.8 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.4 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.1 | 0.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 0.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.6 | GO:0032133 | interphase microtubule organizing center(GO:0031021) chromosome passenger complex(GO:0032133) |
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.9 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 2.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 2.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 2.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 3.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 5.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 3.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.4 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.0 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 3.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 1.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 1.2 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0071953 | fibrinogen complex(GO:0005577) elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0044391 | ribosomal subunit(GO:0044391) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 2.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 1.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 6.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.1 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 5.6 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.6 | 2.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.6 | 1.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.5 | 1.6 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.5 | 3.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.5 | 1.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.5 | 3.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.5 | 1.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.5 | 1.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 2.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 2.6 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.4 | 4.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 1.3 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.4 | 1.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 1.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 2.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.4 | 1.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.4 | 0.7 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.4 | 2.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 1.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 1.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.3 | 1.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 2.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.3 | 2.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.3 | 3.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 2.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.3 | 0.9 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 0.9 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.3 | 0.8 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.3 | 1.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 1.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 2.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 1.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 1.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 1.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 0.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.2 | 2.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 10.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 1.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.6 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.2 | 2.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.0 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 2.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 0.9 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 8.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.5 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.8 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.2 | 2.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 1.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 2.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.5 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 1.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.7 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 2.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 1.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.1 | 0.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.3 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 2.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 4.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 3.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 4.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 1.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 4.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.2 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.8 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 1.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.3 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 1.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.3 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.9 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.2 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.2 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 9.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.2 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.1 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.5 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 2.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 1.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 2.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 1.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 2.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 1.3 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:1990269 | GTP-dependent protein kinase activity(GO:0034211) RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 1.0 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0070736 | protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.5 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.2 | 5.3 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 4.9 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.4 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.7 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 5.5 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 3.0 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 7.4 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.3 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.2 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.7 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.5 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 5.1 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 0.3 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.5 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.7 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.1 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.0 | 2.4 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.4 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 3.8 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.1 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 4.8 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.0 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.3 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.0 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 0.5 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.2 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.3 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.4 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.2 | 2.4 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 8.4 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 5.0 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.1 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 6.7 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 1.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 6.2 | REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 3.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.5 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.7 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.3 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.1 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 13.2 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 1.7 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.8 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 9.3 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.8 | REACTOME_FORMATION_OF_THE_HIV1_EARLY_ELONGATION_COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 1.9 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.3 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.2 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.1 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.5 | REACTOME_SOS_MEDIATED_SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 2.8 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.3 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.9 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 4.9 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.2 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.4 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.8 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.7 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.2 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.5 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.9 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.2 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.9 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.5 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.9 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.9 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.4 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.4 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_THE_MRNA_UPON_BINDING_OF_THE_CAP_BINDING_COMPLEX_AND_EIFS_AND_SUBSEQUENT_BINDING_TO_43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.5 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 2.7 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.0 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.5 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER_AND_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.2 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.7 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.3 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.3 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.9 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 8.1 | REACTOME_CLASS_I_MHC_MEDIATED_ANTIGEN_PROCESSING_PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.0 | 0.3 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 2.2 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME_AQUAPORIN_MEDIATED_TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.1 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.2 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 1.2 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.6 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.0 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.5 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.4 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |





