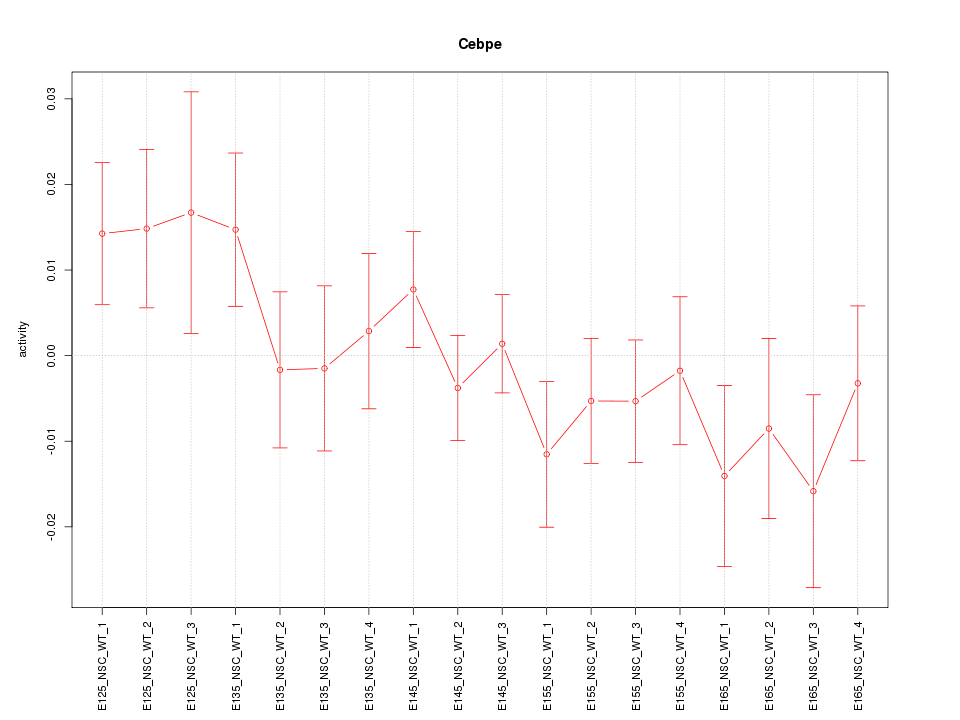
Motif ID: Cebpe
Z-value: 1.028
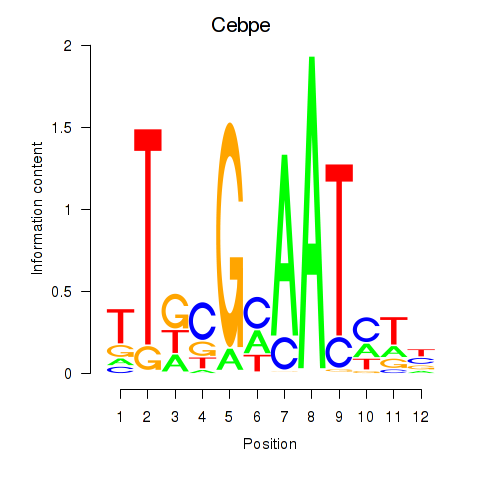
Transcription factors associated with Cebpe:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cebpe | ENSMUSG00000052435.6 | Cebpe |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 1.2 | 6.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.9 | 2.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.7 | 5.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.5 | 1.4 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.4 | 1.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 1.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.4 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.4 | 1.2 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.4 | 1.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.4 | 1.9 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 1.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 1.0 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.3 | 0.6 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.3 | 1.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 1.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 1.3 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 0.8 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.3 | 0.8 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.2 | 3.0 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.2 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.0 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 0.7 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.2 | 1.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.7 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 0.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 0.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.7 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.2 | 1.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 1.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.2 | 0.7 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.6 | GO:2000510 | regulation of MDA-5 signaling pathway(GO:0039533) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 | 0.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 1.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.6 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 0.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 0.4 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 0.6 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.2 | 0.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.2 | 0.5 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.2 | 0.7 | GO:0046655 | dTMP biosynthetic process(GO:0006231) glycine biosynthetic process(GO:0006545) dTMP metabolic process(GO:0046073) folic acid metabolic process(GO:0046655) |
| 0.2 | 1.4 | GO:0015669 | gas transport(GO:0015669) |
| 0.2 | 0.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 0.5 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 0.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 0.6 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.2 | 0.6 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 2.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 0.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 0.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.7 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.7 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.4 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 4.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.4 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.4 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.7 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 1.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:2000812 | response to rapamycin(GO:1901355) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 1.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.6 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.4 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.6 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.1 | 0.5 | GO:0032899 | regulation of neurotrophin production(GO:0032899) positive regulation of neurotrophin production(GO:0032901) |
| 0.1 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.4 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 1.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.7 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 0.6 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.9 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.4 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.3 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.3 | GO:1903225 | regulation of endodermal cell fate specification(GO:0042663) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.2 | GO:1904347 | intestine smooth muscle contraction(GO:0014827) regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) regulation of small intestine smooth muscle contraction(GO:1904347) small intestine smooth muscle contraction(GO:1990770) |
| 0.1 | 0.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.1 | 2.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:1901536 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.1 | 1.2 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.1 | 0.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.2 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.4 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 1.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.3 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.4 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 2.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) positive regulation of interleukin-12 biosynthetic process(GO:0045084) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.4 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 1.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.4 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.3 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.2 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.1 | GO:0000578 | embryonic axis specification(GO:0000578) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.2 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.3 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 1.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.8 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.4 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 2.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.0 | 0.7 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.4 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.5 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.4 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 1.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 1.1 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 1.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 1.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.4 | 1.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.4 | 2.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.3 | 1.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 2.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 0.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 1.2 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 0.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 0.9 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 1.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 3.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 0.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 0.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 1.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.5 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 1.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 1.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 3.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.1 | 1.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.7 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.7 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 0.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 4.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 2.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.1 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.8 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.4 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.5 | 6.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.4 | 2.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 1.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 0.9 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 1.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 1.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.0 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 1.7 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 0.7 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.2 | 0.7 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) serine binding(GO:0070905) |
| 0.2 | 1.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 1.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.4 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.5 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 0.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 0.6 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 0.5 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.2 | 2.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.1 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.7 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 3.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.5 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 1.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.5 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.1 | 1.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.7 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.0 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.1 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.8 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.4 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 3.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.4 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0016835 | carbon-oxygen lyase activity(GO:0016835) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 1.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.4 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 2.9 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 3.4 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.4 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.5 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 5.8 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.7 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.0 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.0 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 3.9 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.5 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.5 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 2.6 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID_GMCSF_PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.2 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 6.0 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 2.6 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 0.6 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.6 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.8 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.2 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 5.4 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.8 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.3 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.1 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 5.0 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.8 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.1 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.5 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.5 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.7 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.8 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.8 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.5 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 5.3 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.0 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.5 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 6.5 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.0 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.4 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.7 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.7 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.9 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME_GLUCAGON_SIGNALING_IN_METABOLIC_REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.5 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME_INHIBITION_OF_THE_PROTEOLYTIC_ACTIVITY_OF_APC_C_REQUIRED_FOR_THE_ONSET_OF_ANAPHASE_BY_MITOTIC_SPINDLE_CHECKPOINT_COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.0 | REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME_PHASE_II_CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.4 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.0 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_SCF_KIT | Genes involved in Signaling by SCF-KIT |