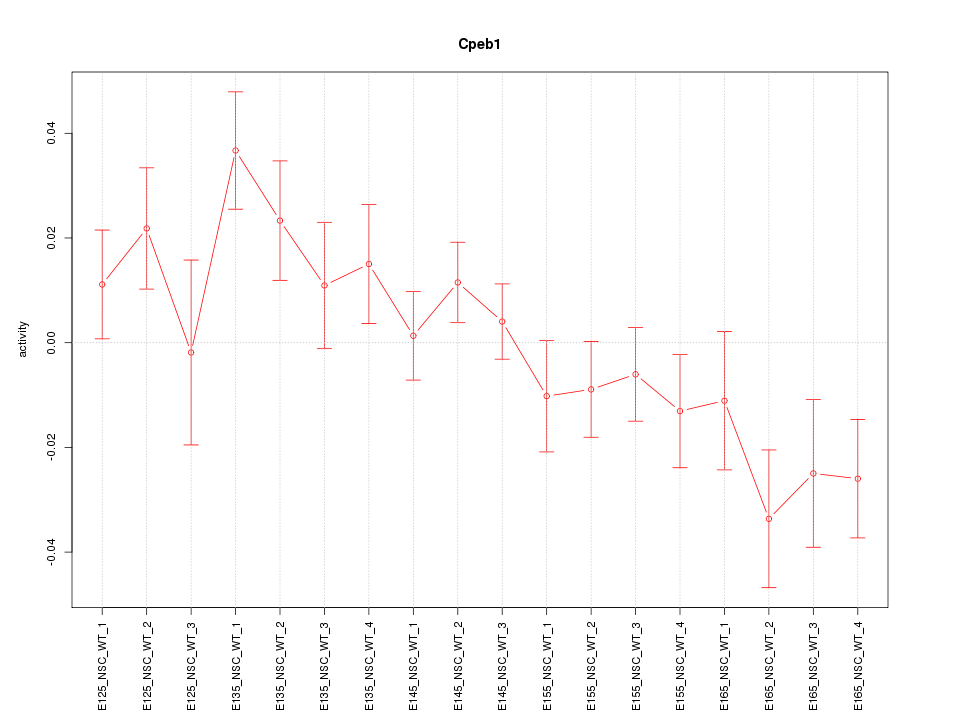
Motif ID: Cpeb1
Z-value: 1.567
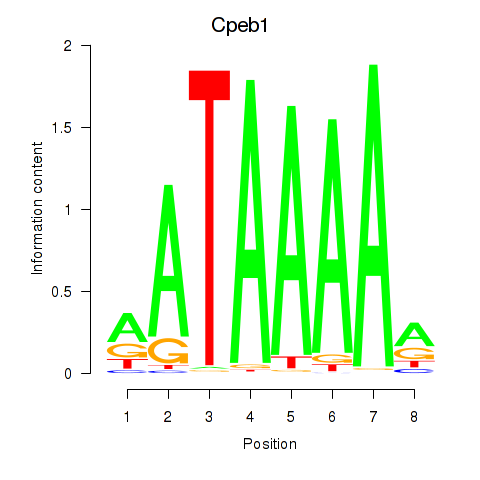
Transcription factors associated with Cpeb1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cpeb1 | ENSMUSG00000025586.10 | Cpeb1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cpeb1 | mm10_v2_chr7_-_81454751_81454764 | -0.35 | 1.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 2.7 | 10.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.9 | 9.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.3 | 6.6 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.2 | 6.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.9 | 2.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.9 | 10.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.8 | 2.5 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.8 | 6.8 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.7 | 2.2 | GO:0071649 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 3.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.6 | 7.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.5 | 1.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.5 | 1.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 1.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.5 | 3.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.5 | 1.4 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.5 | 1.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.4 | 3.6 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.4 | 0.9 | GO:0072199 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.4 | 3.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 1.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 1.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 | 2.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 1.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 1.8 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.4 | 3.2 | GO:0090166 | Golgi disassembly(GO:0090166) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 2.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 1.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 | 1.7 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.3 | 3.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.3 | 1.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 1.3 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.3 | 1.9 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.3 | 1.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 0.9 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.3 | 1.2 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.3 | 1.7 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 3.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 4.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 1.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 1.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 0.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 1.6 | GO:0051604 | protein maturation(GO:0051604) |
| 0.2 | 3.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 0.9 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.2 | 2.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 0.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.6 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.2 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 2.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 1.8 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of receptor catabolic process(GO:2000645) |
| 0.2 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.6 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 1.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 6.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 0.5 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 1.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.5 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.5 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 1.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.6 | GO:2000427 | eosinophil chemotaxis(GO:0048245) T cell extravasation(GO:0072683) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.4 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.4 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 1.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 2.0 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.5 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 4.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.0 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 2.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 1.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 1.4 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 1.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.2 | GO:0035928 | RNA 5'-end processing(GO:0000966) rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.6 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 2.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.7 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 4.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.9 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 3.5 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.4 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.7 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 2.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 | 0.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001267) |
| 0.0 | 1.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.8 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 2.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 5.2 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 1.3 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.2 | GO:2000727 | positive regulation of cardiac muscle cell differentiation(GO:2000727) |
| 0.0 | 0.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0072185 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 1.4 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) negative regulation of neuron maturation(GO:0014043) vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.2 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 1.0 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.8 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 1.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.9 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.2 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.8 | GO:0008623 | CHRAC(GO:0008623) |
| 1.1 | 6.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.8 | 6.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.8 | 6.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.6 | 3.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 2.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 0.6 | GO:0044299 | C-fiber(GO:0044299) |
| 0.3 | 1.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.3 | 1.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 1.5 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 0.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 1.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 3.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 12.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 6.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 14.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 2.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 3.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 11.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 3.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.9 | 9.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.2 | 8.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.1 | 6.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.9 | 5.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 2.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.5 | 1.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.4 | 1.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 1.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 1.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.4 | 1.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 1.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 1.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 1.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 3.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 1.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 2.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.2 | 4.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 4.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 2.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 4.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 0.9 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 2.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 1.7 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.2 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 5.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 14.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 2.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 2.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 12.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.7 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.1 | 0.7 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 1.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 3.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 2.1 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 2.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.0 | 1.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0097371 | BH3 domain binding(GO:0051434) MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 2.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.3 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 3.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.1 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.6 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 10.8 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.0 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.3 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 15.3 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.0 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.8 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.9 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.1 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.1 | 2.4 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.6 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 2.2 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.0 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.6 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.9 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.7 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.2 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 3.5 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.9 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 8.3 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 1.4 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.0 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.0 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.8 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.6 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.7 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.8 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.6 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 2.7 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.6 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.4 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.0 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.3 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.0 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.8 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.4 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.3 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.6 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 3.3 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.0 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.2 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 1.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.2 | REACTOME_CELL_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.8 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.0 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.5 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 5.6 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |