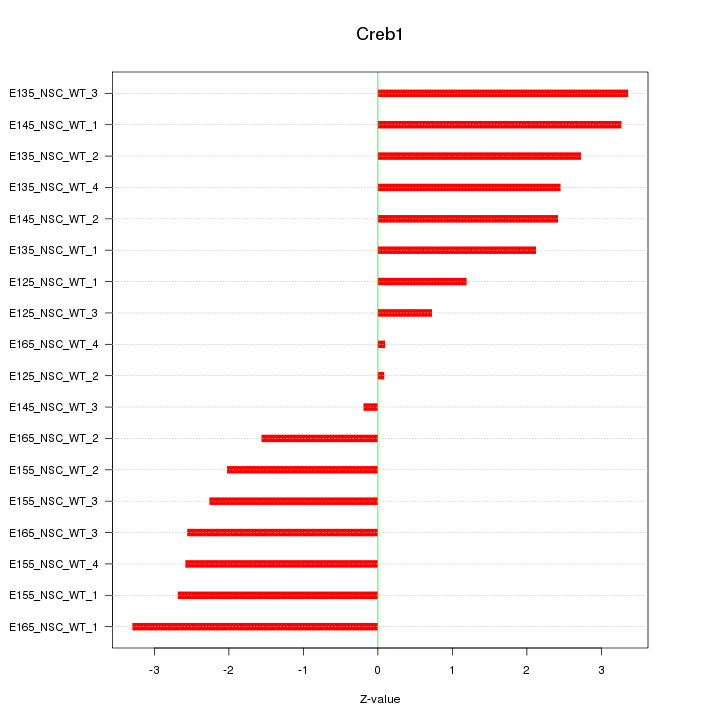
Motif ID: Creb1
Z-value: 2.245
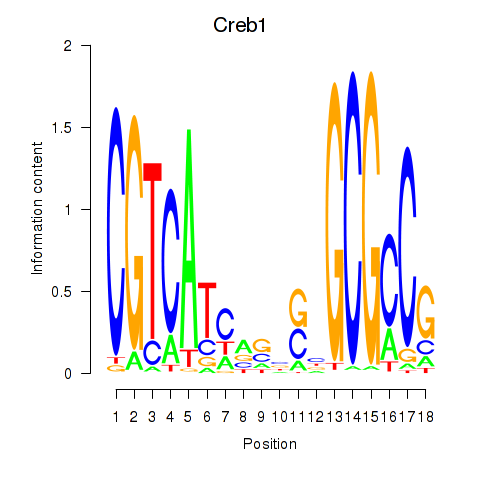
Transcription factors associated with Creb1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Creb1 | ENSMUSG00000025958.8 | Creb1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb1 | mm10_v2_chr1_+_64532790_64532815 | -0.60 | 8.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.1 | 3.4 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.0 | 2.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.9 | 2.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.8 | 2.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.8 | 3.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.8 | 2.3 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.8 | 2.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.7 | 3.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.7 | 2.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.7 | 3.4 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.7 | 2.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.6 | 7.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.6 | 1.8 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.6 | 1.8 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.5 | 1.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 0.5 | 1.4 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) positive regulation of maintenance of sister chromatid cohesion(GO:0034093) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.5 | 2.3 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.5 | 1.9 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.5 | 3.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.4 | 2.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.4 | 2.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 | 2.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 1.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.4 | 1.4 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 1.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.3 | 3.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.3 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.3 | 1.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.3 | 1.0 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.3 | 1.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 1.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 2.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.3 | 1.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 1.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 3.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 1.5 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.3 | 1.0 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 3.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 1.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 3.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 0.7 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.9 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 0.8 | GO:0010796 | regulation of multivesicular body size(GO:0010796) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 2.0 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 2.0 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 0.8 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 1.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 0.6 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.2 | 0.8 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.2 | 0.9 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 2.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.2 | 0.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 2.8 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.7 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.2 | 2.3 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.2 | 0.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 0.5 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 0.6 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.9 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 4.7 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 1.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.5 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 1.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.7 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.9 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 0.5 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 1.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 4.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.5 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.7 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 0.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 1.9 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.1 | 1.6 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 4.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 1.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 5.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.4 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 3.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.3 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.2 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.1 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.6 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 3.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.6 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.0 | 0.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 1.5 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 1.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.5 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 1.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.5 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.1 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) modulation by virus of host autophagy(GO:0039519) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.0 | 0.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 4.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.9 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 1.1 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.6 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 1.0 | 2.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 5.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 3.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.5 | 2.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 1.7 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.4 | 2.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.4 | 2.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 1.5 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.4 | 3.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 1.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 0.9 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.3 | 1.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 3.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.3 | 1.5 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.2 | 2.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.8 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 2.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 1.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 1.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 2.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 6.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 2.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 4.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 6.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 2.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.1 | GO:0097223 | sperm flagellum(GO:0036126) sperm part(GO:0097223) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 2.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.0 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 2.2 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 7.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 4.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 2.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 5.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 2.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 4.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 2.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 2.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.7 | 3.4 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.6 | 1.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 3.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 2.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.4 | 3.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 1.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 1.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 2.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 0.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 2.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 2.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.3 | 1.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.3 | 1.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 1.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 1.5 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.3 | 1.0 | GO:0004844 | mismatch base pair DNA N-glycosylase activity(GO:0000700) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 1.8 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 1.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.7 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.2 | 2.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 5.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 2.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 2.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 6.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 4.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 2.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.2 | 1.8 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 3.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 2.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.8 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 4.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 3.2 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.4 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 2.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 3.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 2.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 4.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.6 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.7 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 2.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 9.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.0 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 5.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 1.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 6.0 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.2 | 1.3 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.1 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.7 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 4.2 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.6 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.3 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.1 | 2.9 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.4 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 3.3 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.3 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.7 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.8 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.8 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID_PLK1_PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 4.3 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 5.1 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 2.3 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 1.5 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 3.5 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.8 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 1.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 0.9 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.1 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.5 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.3 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.5 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.3 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.5 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.8 | REACTOME_ABORTIVE_ELONGATION_OF_HIV1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.2 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.7 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.5 | REACTOME_HOST_INTERACTIONS_OF_HIV_FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 2.2 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.9 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.5 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 8.1 | REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 2.9 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.5 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.4 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 5.6 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 2.5 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.0 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.1 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 2.6 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.0 | 1.0 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.5 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.5 | REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |