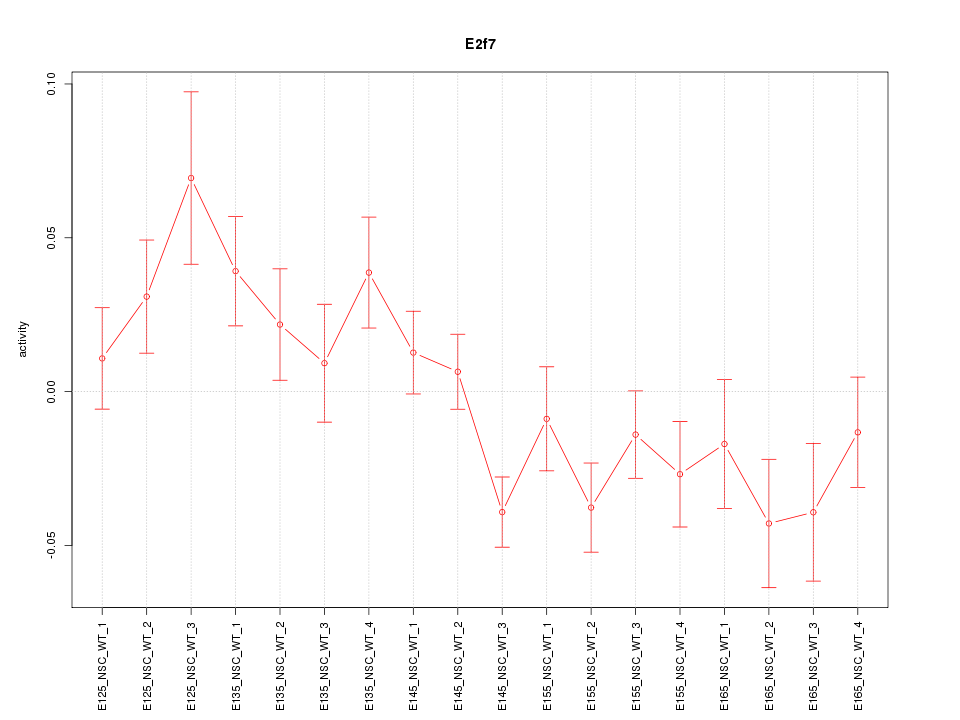
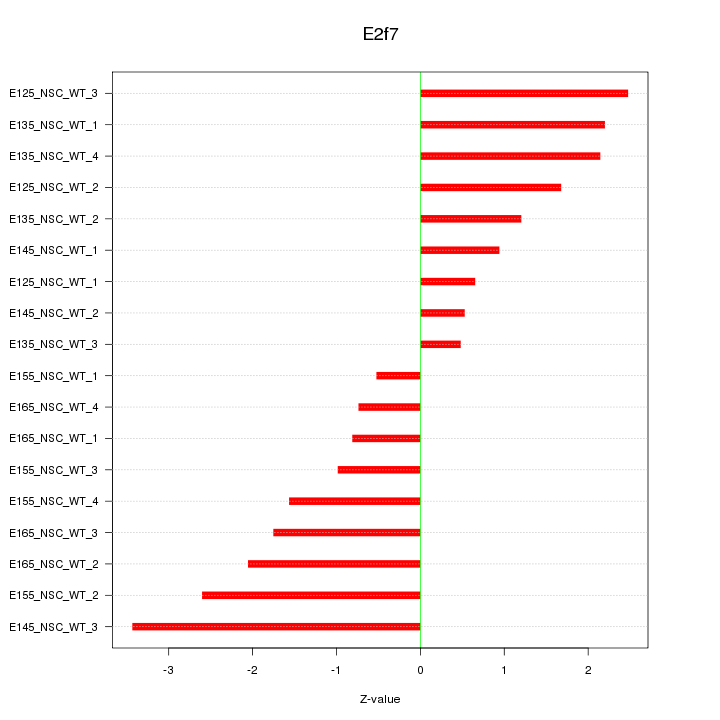
Motif ID: E2f7
Z-value: 1.707
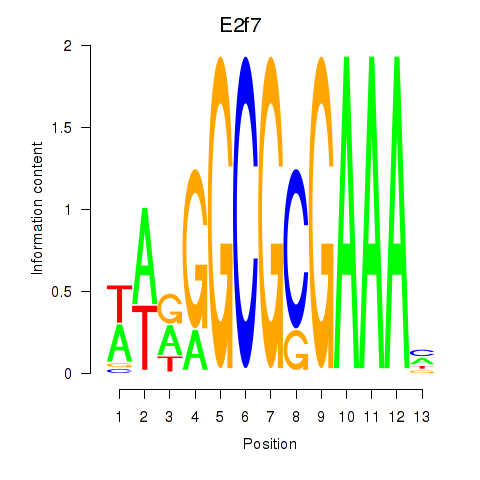
Transcription factors associated with E2f7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f7 | ENSMUSG00000020185.10 | E2f7 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f7 | mm10_v2_chr10_+_110745433_110745572 | 0.48 | 4.3e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.1 | 4.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.6 | 2.8 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.5 | 1.5 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.5 | 2.5 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.5 | 2.5 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.5 | 1.4 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.4 | 7.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 1.3 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.4 | 1.9 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.4 | 3.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 4.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 1.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.0 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.3 | 1.4 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.3 | 1.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 2.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 2.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 0.8 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.3 | 3.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 1.9 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.3 | 2.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 1.0 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 1.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.2 | 0.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 6.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 0.5 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 1.1 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.2 | 0.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 | 2.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 1.2 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.2 | 2.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.6 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 1.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 2.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 10.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 2.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.6 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.1 | 2.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 1.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.0 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 2.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 1.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:1902750 | negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 2.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.8 | 6.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 7.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.6 | 2.5 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.5 | 1.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 1.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 1.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.3 | 4.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 7.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 0.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 7.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 0.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 2.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 4.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 2.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.7 | 8.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.7 | 2.8 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.4 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 2.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 2.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 4.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 1.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 1.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.3 | 0.8 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 3.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 0.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 1.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 1.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 1.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 2.0 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 1.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 4.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 4.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 6.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.6 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 16.2 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.2 | 6.4 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 5.8 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.1 | 7.1 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.4 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.2 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.8 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.6 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.0 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 2.1 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.8 | 7.9 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.5 | 6.3 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.5 | 7.8 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 4.0 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.3 | 11.1 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 4.8 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 5.5 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.2 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.9 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.3 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.6 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.4 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.0 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.0 | 1.0 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |