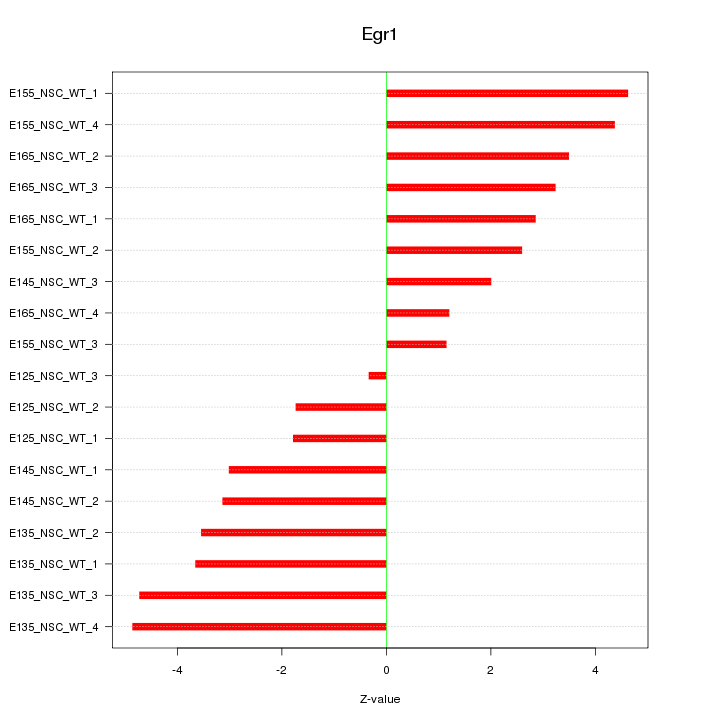
Motif ID: Egr1
Z-value: 3.182
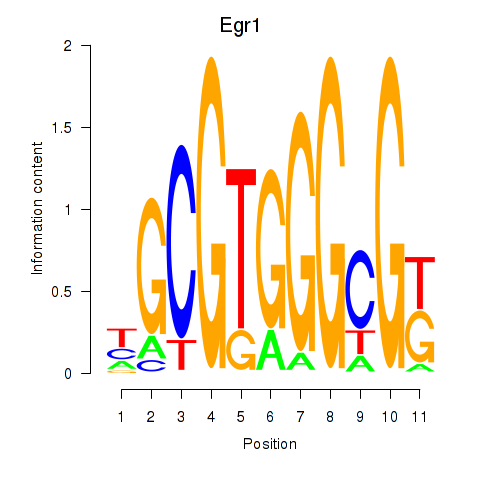
Transcription factors associated with Egr1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Egr1 | ENSMUSG00000038418.7 | Egr1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | mm10_v2_chr18_+_34861200_34861215 | 0.21 | 4.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 35.0 | GO:0032430 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 3.2 | 9.7 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 2.4 | 7.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 2.3 | 9.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 2.2 | 6.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 2.2 | 11.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.9 | 5.7 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 1.7 | 20.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.4 | 4.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.3 | 4.0 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 1.3 | 9.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.2 | 6.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.1 | 6.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.1 | 4.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.1 | 1.1 | GO:0060667 | fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 1.1 | 5.4 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 1.1 | 4.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 1.0 | 3.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.0 | 1.9 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.9 | 4.7 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.9 | 12.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.9 | 4.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.9 | 7.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.9 | 3.6 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.8 | 4.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.8 | 5.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.8 | 3.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.8 | 4.8 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.8 | 3.9 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.8 | 2.3 | GO:0010752 | negative regulation of antigen processing and presentation(GO:0002578) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.8 | 2.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.7 | 11.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 | 1.5 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.7 | 3.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.7 | 3.7 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.7 | 4.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.7 | 9.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.7 | 1.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.7 | 3.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.7 | 4.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.7 | 2.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.7 | 2.0 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.6 | 2.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.6 | 3.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.6 | 3.0 | GO:0090292 | nuclear migration along microfilament(GO:0031022) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.6 | 3.5 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.6 | 2.9 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.6 | 2.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.6 | 5.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.6 | 2.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.5 | 2.2 | GO:0010040 | response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) regulation of peroxidase activity(GO:2000468) |
| 0.5 | 10.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.5 | 2.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.5 | 1.6 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.5 | 2.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.5 | 7.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 3.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.5 | 1.4 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.5 | 1.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 7.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.4 | 2.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.4 | 2.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.4 | 1.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.4 | 5.4 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.4 | 4.1 | GO:0043084 | penile erection(GO:0043084) |
| 0.4 | 0.4 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.4 | 0.8 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 | 1.6 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.4 | 3.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 3.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 1.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) positive regulation of ovulation(GO:0060279) |
| 0.4 | 7.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.4 | 3.0 | GO:0061368 | maternal process involved in parturition(GO:0060137) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.4 | 2.5 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.3 | 3.1 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.3 | 2.4 | GO:0061092 | involuntary skeletal muscle contraction(GO:0003011) detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 1.7 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.3 | 1.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 6.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 1.5 | GO:0015871 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 3.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.2 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.3 | 1.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.3 | 1.5 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.3 | 4.1 | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) |
| 0.3 | 0.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.3 | 1.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 | 1.7 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 1.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 2.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 4.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 0.8 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.3 | 3.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.2 | 1.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 | 3.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.5 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 2.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.2 | 1.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 1.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 4.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.2 | 1.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 2.4 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 2.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 3.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 7.9 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 2.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 1.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 2.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.2 | 0.6 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.2 | 1.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 4.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 0.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 2.9 | GO:0001914 | regulation of T cell mediated cytotoxicity(GO:0001914) |
| 0.2 | 2.5 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 | 1.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 3.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 1.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 3.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.0 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 1.0 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 3.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 5.3 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 0.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.7 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.6 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 2.9 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.9 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 3.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.3 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 3.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 5.5 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 2.0 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 1.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 3.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.2 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 3.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.8 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.5 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 0.6 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.5 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 9.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 3.3 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.1 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 0.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 5.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 3.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.7 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.6 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 1.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 2.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 2.9 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 1.8 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 0.9 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 1.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.8 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 8.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.9 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.6 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.9 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 2.2 | GO:1900076 | regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.0 | 1.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 4.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 2.6 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.6 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.4 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 3.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 3.3 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 1.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.0 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.7 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.6 | GO:0042770 | signal transduction in response to DNA damage(GO:0042770) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.8 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.8 | GO:0090537 | CERF complex(GO:0090537) |
| 1.7 | 10.1 | GO:0008091 | spectrin(GO:0008091) |
| 1.6 | 9.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.5 | 20.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.4 | 5.7 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.2 | 3.7 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.1 | 6.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.0 | 33.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.9 | 2.7 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.9 | 6.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.8 | 3.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.8 | 3.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.8 | 7.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.8 | 2.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 1.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.6 | 8.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.5 | 1.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.5 | 7.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 5.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 5.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.4 | 4.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 3.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.4 | 10.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 3.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 1.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.4 | 17.5 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.4 | 6.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 2.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 1.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.3 | 8.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 0.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 17.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 2.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 3.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.1 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.2 | 2.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 5.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 8.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 2.9 | GO:0031105 | septin complex(GO:0031105) |
| 0.2 | 2.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.7 | GO:0043293 | apoptosome(GO:0043293) |
| 0.2 | 2.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 5.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.1 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 12.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 3.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 4.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 3.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 1.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 17.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.2 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 3.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.8 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 4.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 7.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 10.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 3.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 5.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 4.1 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 2.4 | 7.3 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 2.3 | 9.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.8 | 20.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.8 | 5.4 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 1.4 | 4.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.4 | 36.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.2 | 5.0 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 1.2 | 11.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 1.1 | 15.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.1 | 4.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.0 | 6.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.0 | 5.7 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.9 | 2.8 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.9 | 3.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.9 | 3.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.9 | 5.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 3.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.8 | 10.9 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.8 | 4.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.8 | 2.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.8 | 3.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.8 | 2.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 3.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.7 | 12.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.7 | 7.8 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.7 | 9.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 2.0 | GO:0019966 | C-X-C chemokine binding(GO:0019958) interleukin-1 binding(GO:0019966) tumor necrosis factor binding(GO:0043120) |
| 0.6 | 1.8 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.6 | 1.8 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.6 | 1.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.6 | 4.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 2.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 1.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 1.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 1.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.4 | 2.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 6.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.4 | 3.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.4 | 4.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 2.6 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.4 | 2.9 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.4 | 2.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 1.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.3 | 1.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.3 | 1.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.7 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 1.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 3.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 3.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 2.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 1.8 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 6.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 6.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 5.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 0.7 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 4.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 2.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 6.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 0.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.8 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 1.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 4.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 2.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 3.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 6.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 3.6 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 3.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 6.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.4 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 4.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 11.4 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 5.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 4.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 3.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 5.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 6.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 0.7 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 3.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 5.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 6.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 2.5 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 7.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 5.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.2 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 4.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.7 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.6 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 33.1 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.6 | 8.8 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 22.3 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.4 | 0.9 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 13.7 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 7.8 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 5.4 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 4.8 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.2 | 9.7 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 6.3 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.2 | 9.8 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.2 | 2.1 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.7 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 3.2 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 2.7 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.4 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 4.9 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 8.3 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 4.7 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.6 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.2 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.2 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.4 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.7 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.4 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.1 | 0.6 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 2.5 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.9 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.8 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.1 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.7 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 4.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 40.8 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 1.6 | 11.3 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.9 | 20.5 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.8 | 9.1 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.7 | 8.1 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.7 | 9.8 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.5 | 4.3 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 12.5 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 17.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 3.8 | REACTOME_DAG_AND_IP3_SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.4 | 4.2 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 8.3 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.3 | 1.1 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 2.2 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 2.2 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.2 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 3.4 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 5.0 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.2 | 2.0 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 1.8 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.9 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 2.8 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 5.5 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 6.2 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 3.1 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.3 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 4.0 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 10.4 | REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 1.1 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.2 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 5.0 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 0.8 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.7 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.9 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.0 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.7 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 0.4 | REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 1.3 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 7.3 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.8 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 1.1 | REACTOME_CTLA4_INHIBITORY_SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.5 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.1 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.3 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 9.3 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.3 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.0 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.5 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |