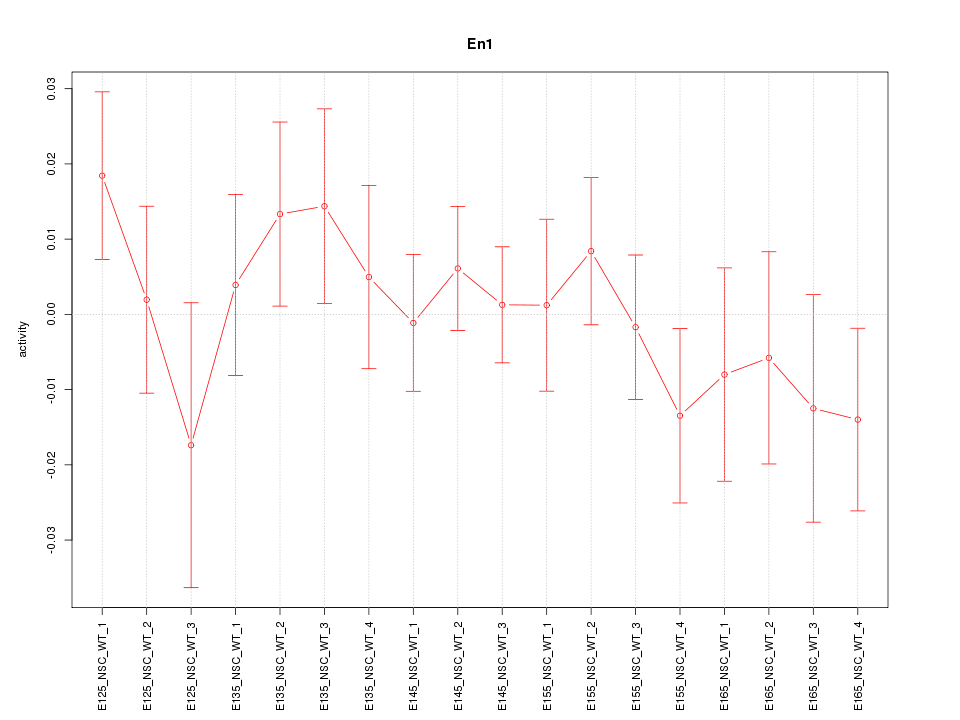
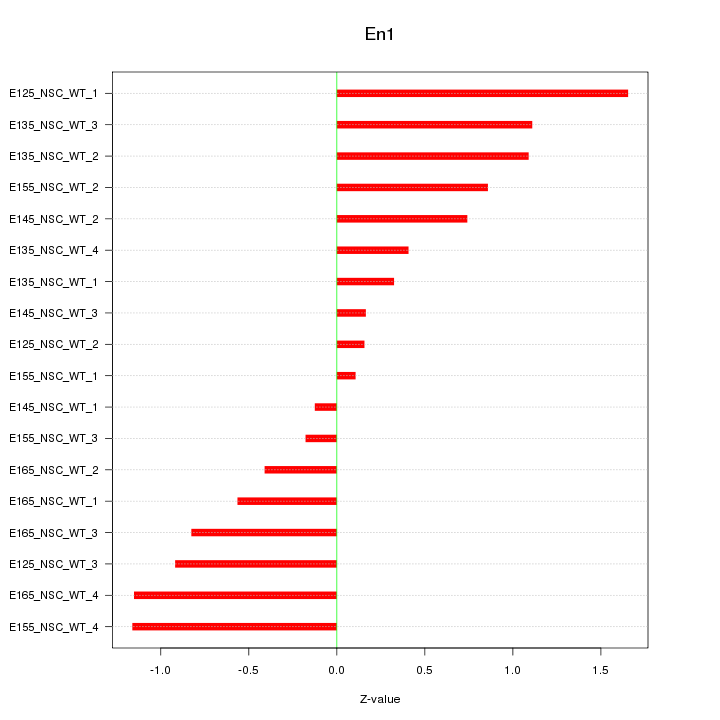
Motif ID: En1
Z-value: 0.800
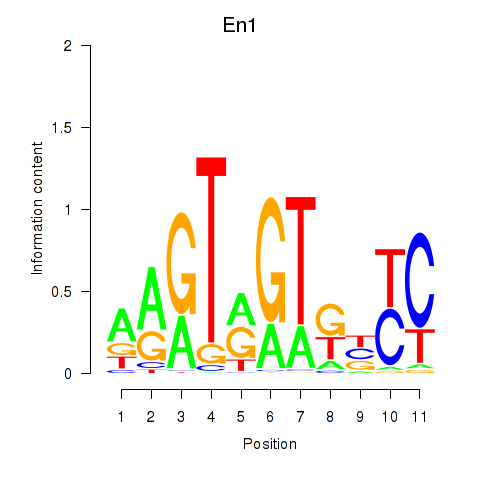
Transcription factors associated with En1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| En1 | ENSMUSG00000058665.7 | En1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En1 | mm10_v2_chr1_+_120602405_120602418 | -0.49 | 3.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.3 | 0.9 | GO:0003195 | tricuspid valve formation(GO:0003195) |
| 0.3 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 0.7 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 | 1.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 0.6 | GO:0097278 | transforming growth factor beta activation(GO:0036363) complement-dependent cytotoxicity(GO:0097278) |
| 0.2 | 1.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 0.6 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 0.6 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) negative regulation of intestinal absorption(GO:1904479) |
| 0.2 | 0.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.5 | GO:0042713 | operant conditioning(GO:0035106) sperm ejaculation(GO:0042713) |
| 0.2 | 1.0 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 0.5 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 2.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.5 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.1 | 0.9 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.5 | GO:0060558 | regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.1 | 0.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.4 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.6 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.4 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 0.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.7 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.1 | 0.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 | 0.4 | GO:0019659 | fermentation(GO:0006113) lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.8 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.8 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.1 | 0.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.7 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.1 | 1.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.2 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) GPI anchor release(GO:0006507) |
| 0.1 | 0.4 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.2 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.1 | 0.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 0.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.1 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.1 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.6 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.0 | 0.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.3 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.0 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.3 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.0 | 0.5 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.6 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 1.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.7 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.8 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.0 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 0.8 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.2 | 1.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.5 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.3 | 0.9 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 1.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 0.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.7 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 1.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.6 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.7 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 1.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.4 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.7 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.4 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.4 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.9 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.1 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 1.7 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.4 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.4 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.6 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.3 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.4 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.2 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.8 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.6 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME_NRIF_SIGNALS_CELL_DEATH_FROM_THE_NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.4 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.4 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.6 | REACTOME_LIPID_DIGESTION_MOBILIZATION_AND_TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.4 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |