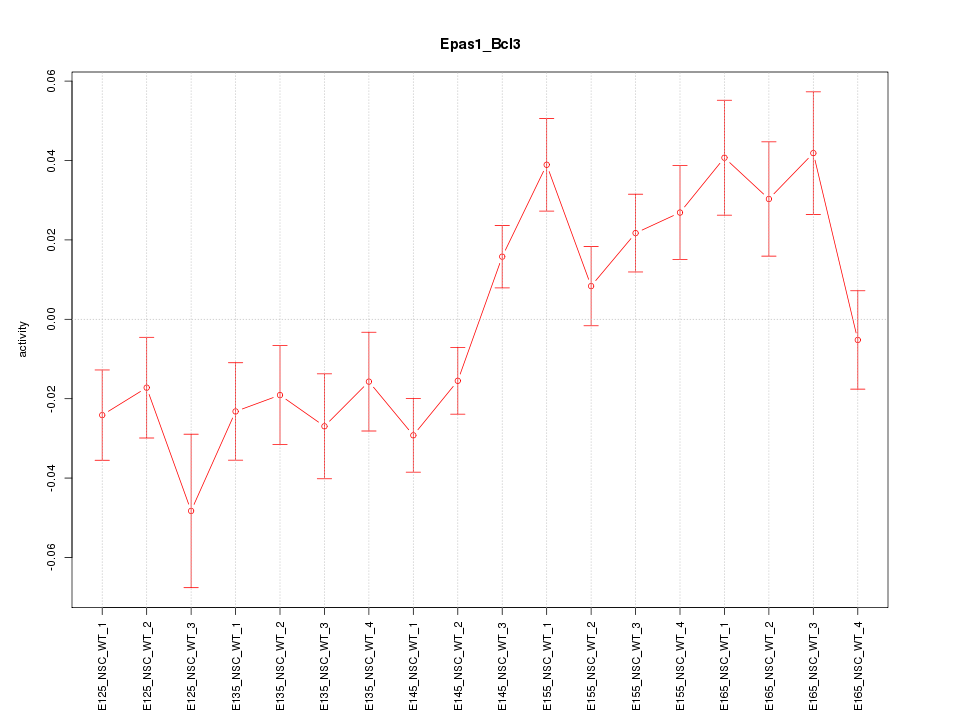
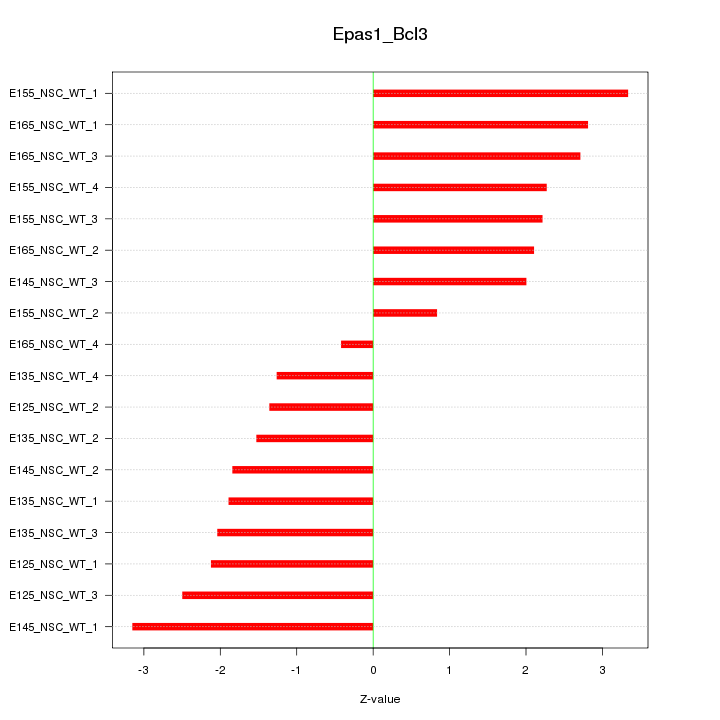
Motif ID: Epas1_Bcl3
Z-value: 2.152
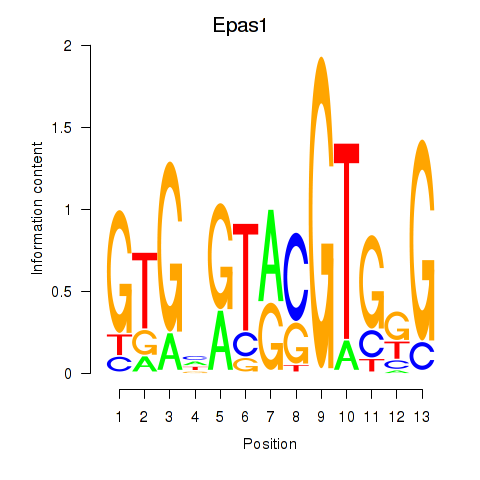
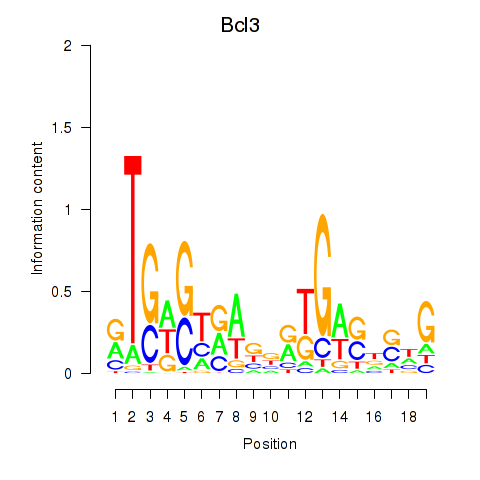
Transcription factors associated with Epas1_Bcl3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bcl3 | ENSMUSG00000053175.10 | Bcl3 |
| Epas1 | ENSMUSG00000024140.9 | Epas1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Epas1 | mm10_v2_chr17_+_86753900_86753914 | 0.69 | 1.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 2.7 | 10.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.9 | 13.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 1.6 | 12.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.5 | 6.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.4 | 8.2 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 1.2 | 7.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 1.0 | 3.1 | GO:0098528 | terpenoid catabolic process(GO:0016115) skeletal muscle fiber differentiation(GO:0098528) |
| 0.9 | 5.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.9 | 4.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.8 | 7.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.8 | 7.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.8 | 3.8 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.7 | 2.0 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.6 | 3.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.6 | 3.7 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.6 | 1.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.6 | 3.9 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.6 | 2.8 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.5 | 2.7 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.5 | 2.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.5 | 1.0 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.5 | 2.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.5 | 2.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 1.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 6.9 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.5 | 1.4 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.4 | 3.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.4 | 0.9 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.4 | 1.3 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 1.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.4 | 2.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.4 | 1.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.4 | 3.6 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.4 | 0.7 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.4 | 1.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 0.7 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.3 | 2.7 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 1.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 | 1.7 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.3 | 3.6 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.3 | 1.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 1.0 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.3 | 1.9 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.3 | 0.9 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.3 | 4.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.3 | 8.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 3.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 1.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 4.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 1.2 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.2 | 4.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 1.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 1.4 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 0.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 2.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 4.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.8 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.2 | 0.6 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 | 1.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 0.2 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.2 | 5.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 9.6 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.2 | 8.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 0.7 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 3.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 3.4 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.2 | 0.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 1.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 1.6 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 | 0.8 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 0.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 1.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.7 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 1.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 1.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 1.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 4.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 2.0 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 2.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 1.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.3 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.1 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.8 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 1.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 3.1 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 2.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.8 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 3.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 8.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.2 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 1.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 2.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.8 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.1 | 3.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 3.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.4 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 1.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 | 1.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 1.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 1.4 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.1 | 3.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:1903756 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 3.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:1904706 | detection of oxygen(GO:0003032) negative regulation of macrophage cytokine production(GO:0010936) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 3.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 2.1 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 1.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 3.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 1.1 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.4 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0043512 | inhibin A complex(GO:0043512) |
| 1.3 | 6.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.1 | 4.5 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.9 | 4.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.9 | 5.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 15.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.7 | 2.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.6 | 2.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.6 | 1.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.5 | 4.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 7.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 1.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 1.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 1.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 7.1 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 3.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 2.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.3 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 2.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 2.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 2.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 5.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 2.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 2.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.2 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 0.9 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 4.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 7.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 5.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 6.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 4.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 8.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 5.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 19.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 6.1 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 6.7 | GO:0097458 | neuron part(GO:0097458) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 2.1 | 6.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.8 | 10.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.5 | 6.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.9 | 8.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.9 | 2.7 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.9 | 4.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.8 | 2.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.8 | 4.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.6 | 4.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.6 | 3.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.6 | 3.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.6 | 2.3 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.6 | 3.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 2.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 3.8 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.5 | 13.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.5 | 1.4 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 0.4 | 1.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 1.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.4 | 1.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.4 | 4.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.4 | 1.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 2.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 3.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 1.3 | GO:0098988 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled glutamate receptor activity(GO:0098988) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.3 | 11.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 2.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 1.5 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.3 | 2.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 0.9 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.3 | 1.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 3.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 13.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 3.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 2.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 2.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 2.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 5.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 1.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 4.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 2.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 3.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.2 | 6.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.7 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 3.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 0.7 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 0.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 2.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.5 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.2 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 0.9 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.1 | 1.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 7.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 4.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 2.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.0 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 2.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.9 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 0.7 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 0.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 3.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 4.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 5.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 1.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) primary miRNA binding(GO:0070878) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.9 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 3.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 5.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 5.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 9.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 3.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.5 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.4 | 11.0 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.2 | 10.0 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.2 | 7.4 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.2 | 1.7 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.7 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.1 | 8.5 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 3.6 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 0.4 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.0 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.7 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.7 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.1 | 3.8 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.8 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.0 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.4 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.9 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 1.0 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 3.2 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 8.2 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 11.1 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 13.2 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 4.3 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 4.5 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.7 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 3.1 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 8.1 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 3.9 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.2 | 7.7 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 1.7 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.9 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 2.2 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.0 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 2.2 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 5.1 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 4.1 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 5.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.1 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.7 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 8.1 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.4 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.9 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.1 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 0.5 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.2 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.1 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.8 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.4 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 6.5 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.3 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.7 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.8 | REACTOME_HEPARAN_SULFATE_HEPARIN_HS_GAG_METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.0 | 1.4 | REACTOME_NGF_SIGNALLING_VIA_TRKA_FROM_THE_PLASMA_MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.9 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |