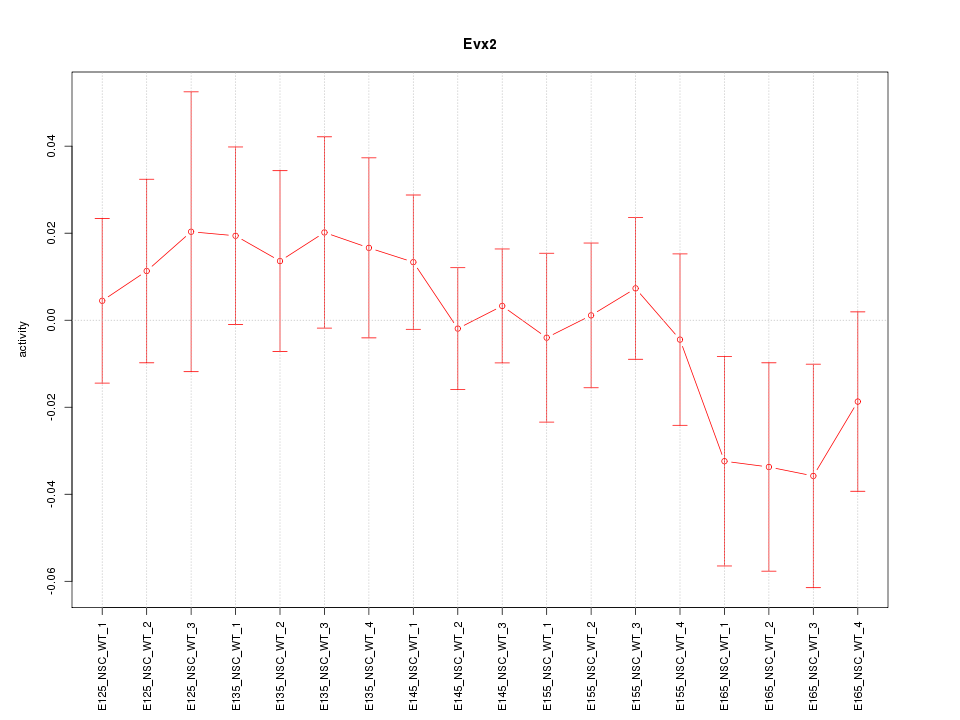
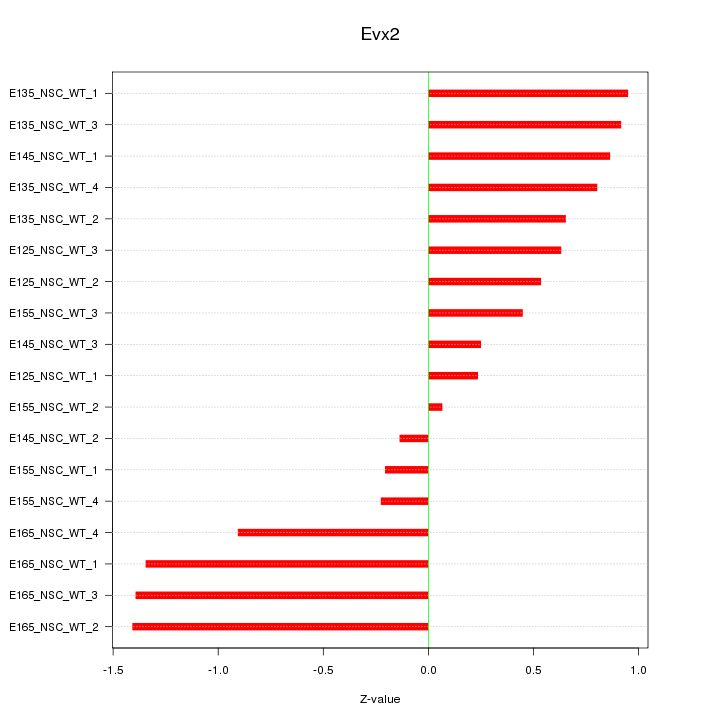
Motif ID: Evx2
Z-value: 0.791
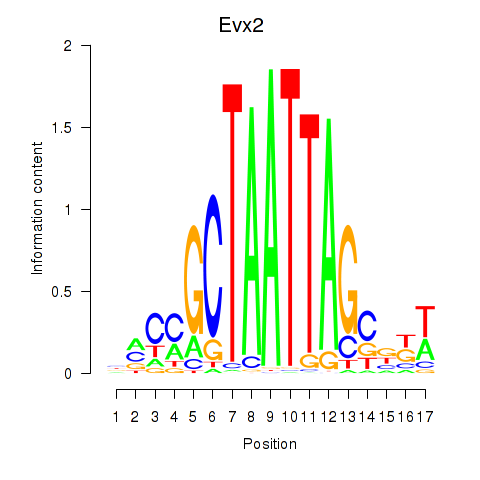
Transcription factors associated with Evx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Evx2 | ENSMUSG00000001815.9 | Evx2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.5 | 1.4 | GO:0009826 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 7.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 1.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.3 | 0.8 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 4.8 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 1.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 2.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 1.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.8 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 3.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 1.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 8.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 3.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.4 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 1.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 4.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 13.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.8 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 3.3 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 1.3 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.2 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 4.3 | REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 1.2 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |