Motif ID: Foxd2
Z-value: 1.236
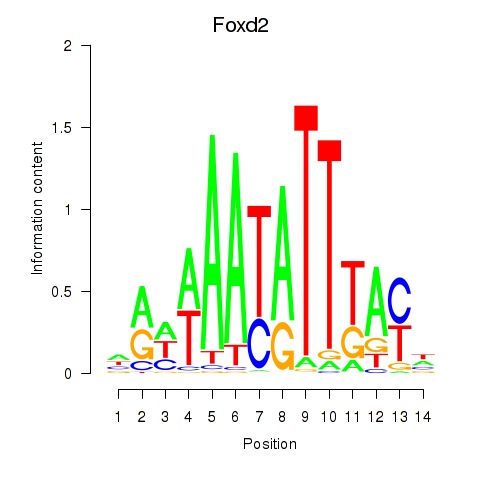
Transcription factors associated with Foxd2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxd2 | ENSMUSG00000055210.3 | Foxd2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.5 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.1 | 4.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.7 | 4.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.5 | 2.5 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.5 | 2.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.4 | 2.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 2.3 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 | 2.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 0.9 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 0.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 2.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 1.4 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 | 1.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.9 | GO:0042228 | interleukin-8 biosynthetic process(GO:0042228) |
| 0.2 | 0.5 | GO:2001180 | regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 1.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 6.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 1.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 2.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 1.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.3 | GO:0006547 | histidine metabolic process(GO:0006547) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 3.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 9.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.6 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 2.5 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.6 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 2.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 3.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.0 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.5 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.8 | 2.5 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.5 | 4.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 3.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 0.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 2.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 2.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 7.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 6.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 1.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 7.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 2.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 9.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.2 | 4.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.6 | 2.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 4.6 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.4 | 2.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 2.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 4.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 2.8 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 6.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 0.6 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 7.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 2.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 6.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.5 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 7.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.1 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.4 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.1 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.2 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.0 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.2 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.1 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.4 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 4.6 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.2 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 4.1 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.4 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 2.3 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME_TRAF6_MEDIATED_INDUCTION_OF_TAK1_COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.6 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |





