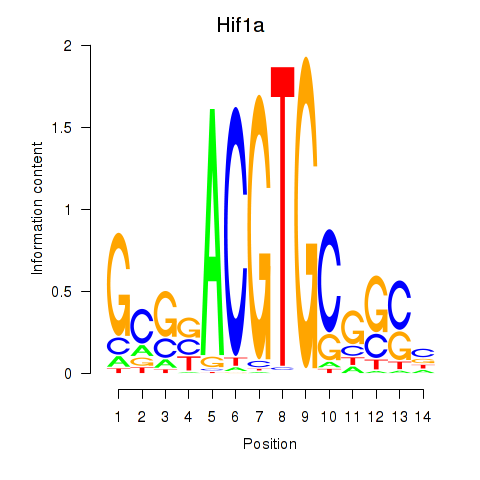
Motif ID: Hif1a
Z-value: 0.902
Transcription factors associated with Hif1a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hif1a | ENSMUSG00000021109.7 | Hif1a |
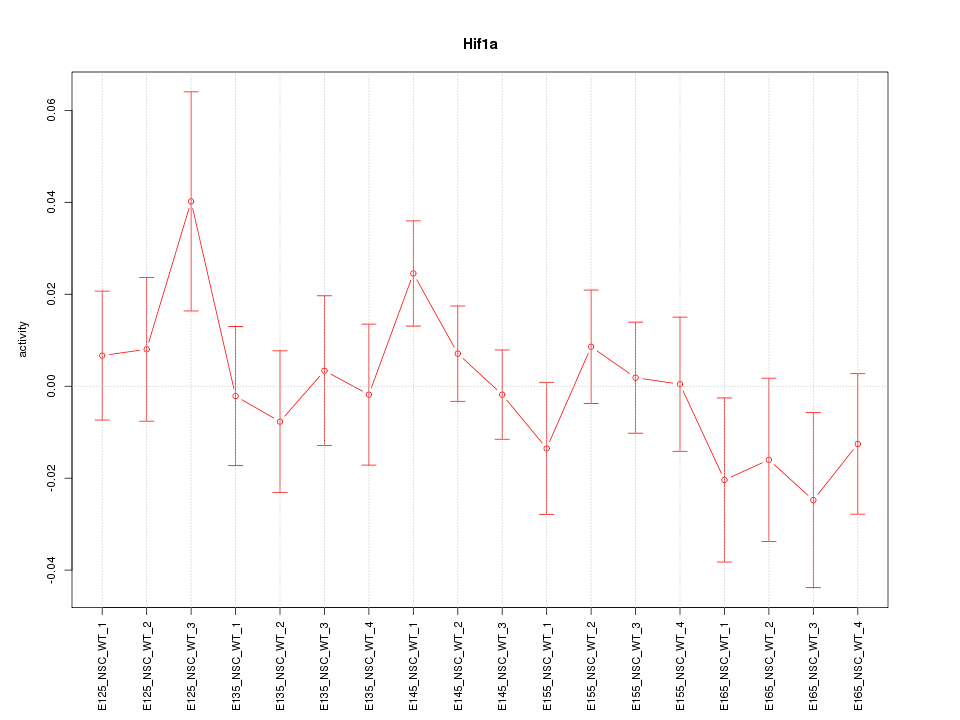
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hif1a | mm10_v2_chr12_+_73907904_73907970 | -0.63 | 5.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.9 | 2.7 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.7 | 2.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 5.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.7 | 0.7 | GO:0033122 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.7 | 2.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.6 | 2.5 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.5 | 2.7 | GO:0019659 | fermentation(GO:0006113) lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.5 | 1.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.5 | 1.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.5 | 1.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.4 | 1.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 | 1.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.3 | 1.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 0.9 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 2.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 0.9 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.3 | 3.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.3 | 0.8 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 1.0 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.3 | 0.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 1.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 0.7 | GO:0007621 | negative regulation of female receptivity(GO:0007621) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of cellular pH reduction(GO:0032847) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.7 | GO:0010248 | B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 1.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 1.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 0.6 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 0.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.4 | GO:0097278 | transforming growth factor beta activation(GO:0036363) complement-dependent cytotoxicity(GO:0097278) |
| 0.1 | 0.7 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.4 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.3 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) response to cobalt ion(GO:0032025) mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.3 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 1.1 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.7 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 4.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.3 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.1 | 0.2 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.3 | GO:0009052 | D-ribose metabolic process(GO:0006014) pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.4 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.5 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 2.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 0.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.3 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 1.0 | GO:0046475 | bleb assembly(GO:0032060) hydrogen peroxide catabolic process(GO:0042744) glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.3 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.7 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0033522 | histone H2A ubiquitination(GO:0033522) histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 2.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.2 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 1.2 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.7 | GO:0010470 | regulation of gastrulation(GO:0010470) |
| 0.0 | 0.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 1.2 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.2 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.4 | GO:0007129 | synapsis(GO:0007129) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 2.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 1.7 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.4 | 2.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 1.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 5.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 2.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 1.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.9 | 2.7 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.7 | 2.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 1.5 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.5 | 1.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 2.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.4 | 1.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 5.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 1.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.3 | 1.2 | GO:0030292 | hyalurononglucosaminidase activity(GO:0004415) protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 4.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 1.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.7 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 0.7 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 0.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 2.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.2 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.4 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.9 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 1.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 2.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 2.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.8 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 1.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.5 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.2 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.3 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.5 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 2.2 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.2 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 2.7 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.0 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID_AURORA_A_PATHWAY | Aurora A signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.7 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 11.0 | REACTOME_GLUCOSE_METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 2.1 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.2 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.0 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.3 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.1 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.7 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.2 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.2 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.4 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.2 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 4.1 | REACTOME_ACTIVATION_OF_THE_MRNA_UPON_BINDING_OF_THE_CAP_BINDING_COMPLEX_AND_EIFS_AND_SUBSEQUENT_BINDING_TO_43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 1.3 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.2 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.0 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.2 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.6 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.7 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.1 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |