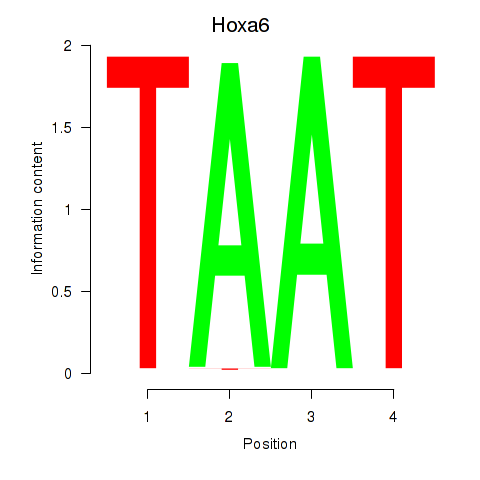
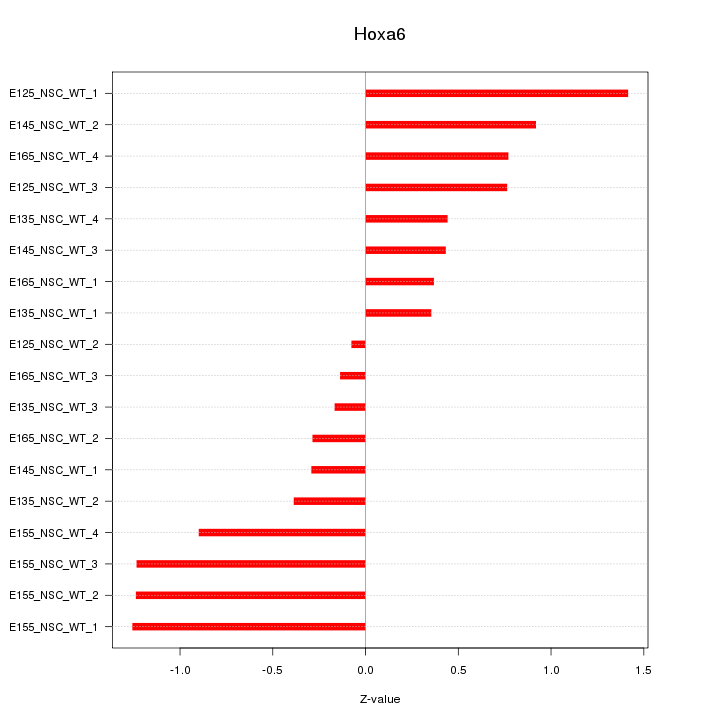
| 0.9 |
3.7 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.5 |
1.6 |
GO:0048389 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) negative regulation of immature T cell proliferation in thymus(GO:0033088) cloaca development(GO:0035844) intermediate mesoderm development(GO:0048389) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) pattern specification involved in mesonephros development(GO:0061227) regulation of hepatocyte differentiation(GO:0070366) anterior/posterior pattern specification involved in kidney development(GO:0072098) negative regulation of glomerular mesangial cell proliferation(GO:0072125) metanephric S-shaped body morphogenesis(GO:0072284) negative regulation of glomerulus development(GO:0090194) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) cardiac jelly development(GO:1905072) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.5 |
5.8 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 |
3.2 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.4 |
1.2 |
GO:0070172 |
oculomotor nerve development(GO:0021557) positive regulation of tooth mineralization(GO:0070172) |
| 0.3 |
1.0 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) negative regulation of keratinocyte differentiation(GO:0045617) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.3 |
0.9 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.3 |
1.1 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.3 |
1.9 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 |
0.8 |
GO:1900108 |
negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 |
2.1 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.2 |
0.7 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.2 |
0.7 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 |
2.6 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.2 |
0.5 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.2 |
0.5 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.2 |
1.5 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.2 |
0.3 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.1 |
0.4 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 |
0.6 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.3 |
GO:2000675 |
negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 |
1.9 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
0.5 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.5 |
GO:0060022 |
hard palate development(GO:0060022) |
| 0.1 |
1.5 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
1.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.7 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
0.4 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.1 |
0.2 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.6 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.5 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 |
0.4 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 |
0.4 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.9 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 |
0.5 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.2 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.6 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.9 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.3 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.2 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 |
0.3 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 |
0.2 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 |
1.0 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.0 |
1.0 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.3 |
GO:0015791 |
polyol transport(GO:0015791) bile acid secretion(GO:0032782) |
| 0.0 |
0.1 |
GO:1900141 |
regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) |
| 0.0 |
0.4 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.0 |
0.3 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.8 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.9 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 |
0.2 |
GO:0033484 |
nitric oxide homeostasis(GO:0033484) |
| 0.0 |
0.1 |
GO:0070494 |
regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 |
1.2 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.2 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.1 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 |
0.8 |
GO:0010259 |
multicellular organism aging(GO:0010259) |
| 0.0 |
0.3 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
1.9 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.1 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.5 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.7 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.7 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.3 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.5 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.1 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 |
0.1 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.1 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.3 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.3 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.2 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 |
0.1 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.3 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.1 |
GO:0060849 |
regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |