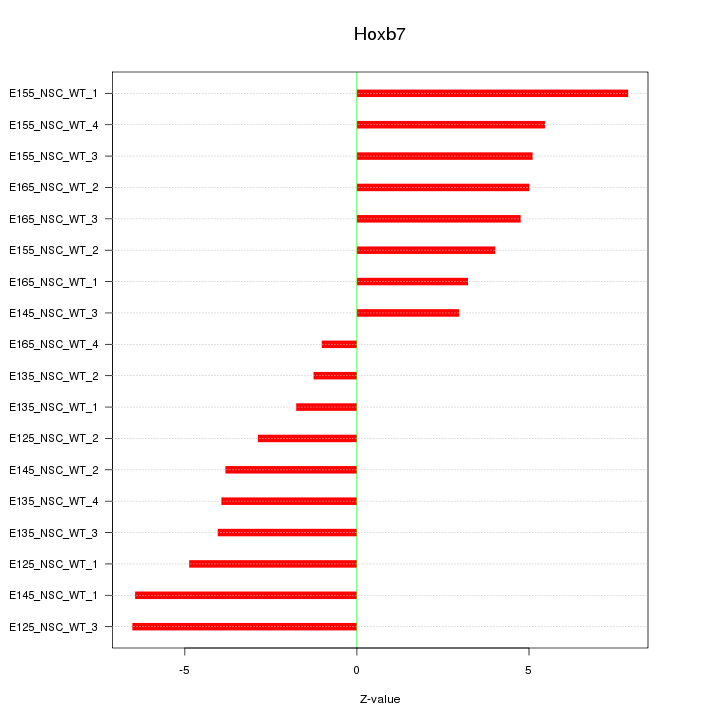
Motif ID: Hoxb7
Z-value: 4.531
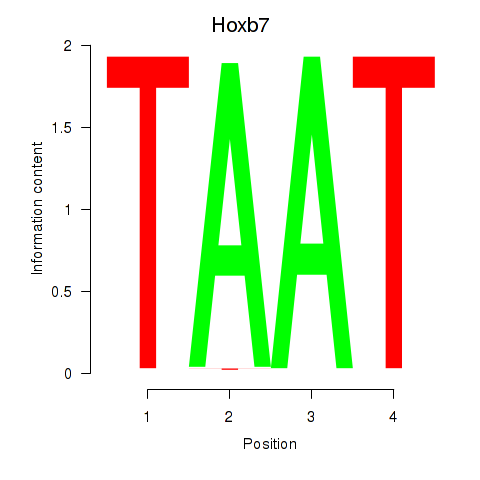
Transcription factors associated with Hoxb7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxb7 | ENSMUSG00000038721.8 | Hoxb7 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 43.0 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 10.1 | 30.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 8.6 | 25.8 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 5.8 | 17.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 5.7 | 51.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 5.5 | 21.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 5.3 | 31.8 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 5.1 | 15.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 4.8 | 62.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 4.0 | 19.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 3.6 | 40.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 3.5 | 17.3 | GO:0042701 | progesterone secretion(GO:0042701) |
| 3.4 | 13.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 3.3 | 26.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 3.2 | 16.0 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 3.2 | 12.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.9 | 26.5 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 2.6 | 7.9 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 2.5 | 35.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 2.5 | 7.6 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 2.5 | 10.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 2.5 | 12.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 2.4 | 17.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.4 | 4.8 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 2.4 | 16.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 2.4 | 2.4 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 2.4 | 23.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 2.3 | 28.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 2.2 | 6.7 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 2.2 | 15.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 2.2 | 8.8 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 2.2 | 28.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 2.1 | 6.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 2.1 | 34.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 2.1 | 8.5 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 2.1 | 4.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 2.1 | 86.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 2.1 | 8.2 | GO:2000795 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 2.0 | 12.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 2.0 | 8.2 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.0 | 17.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.9 | 15.4 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 1.9 | 5.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.8 | 5.4 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.7 | 10.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 1.6 | 13.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.6 | 3.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.5 | 9.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 1.5 | 4.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 1.5 | 7.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 1.5 | 7.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.4 | 58.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 1.4 | 4.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.4 | 23.0 | GO:0097090 | presynaptic membrane organization(GO:0097090) |
| 1.3 | 20.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.3 | 4.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 1.3 | 3.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.3 | 3.9 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 1.3 | 9.0 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 1.3 | 6.4 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.3 | 6.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.3 | 2.5 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.3 | 5.0 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.2 | 3.7 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.2 | 20.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 1.2 | 7.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.2 | 9.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 1.1 | 14.5 | GO:0001553 | luteinization(GO:0001553) |
| 1.1 | 8.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.1 | 5.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.1 | 3.3 | GO:1904978 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 1.1 | 1.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 1.1 | 5.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.1 | 8.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.0 | 8.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.0 | 3.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 1.0 | 3.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 1.0 | 11.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 1.0 | 15.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.0 | 2.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 1.0 | 2.0 | GO:0071872 | response to epinephrine(GO:0071871) cellular response to epinephrine stimulus(GO:0071872) |
| 1.0 | 14.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.0 | 3.9 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.9 | 5.5 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.9 | 3.7 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.9 | 37.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.9 | 4.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.9 | 6.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.9 | 18.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.9 | 10.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.8 | 2.5 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.8 | 4.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.8 | 2.5 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.8 | 4.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.8 | 2.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 6.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.8 | 5.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.8 | 21.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.8 | 5.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.8 | 4.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 2.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.8 | 3.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.8 | 2.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.8 | 12.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.8 | 2.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.7 | 6.0 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.7 | 10.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.7 | 5.1 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.7 | 2.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 7.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.7 | 2.2 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.7 | 2.9 | GO:0072362 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.7 | 3.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.7 | 2.1 | GO:0003289 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) noradrenergic neuron differentiation(GO:0003357) |
| 0.7 | 4.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.7 | 2.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.7 | 2.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.7 | 3.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.7 | 2.0 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.7 | 16.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.7 | 2.7 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.7 | 3.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 3.9 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.6 | 1.9 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.6 | 1.8 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.6 | 2.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.6 | 3.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.6 | 2.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.6 | 1.8 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.6 | 2.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.6 | 3.0 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.6 | 1.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.6 | 8.8 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.6 | 2.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.6 | 1.7 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.6 | 4.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.6 | 2.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.6 | 1.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.6 | 12.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.6 | 2.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.6 | 5.0 | GO:0060613 | fat pad development(GO:0060613) |
| 0.5 | 4.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.5 | 2.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.5 | 1.6 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.5 | 1.6 | GO:2000016 | regulation of determination of dorsal identity(GO:2000015) negative regulation of determination of dorsal identity(GO:2000016) |
| 0.5 | 2.6 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.5 | 15.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.5 | 3.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.5 | 1.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.5 | 23.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.5 | 10.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.5 | 3.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 3.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 2.0 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.5 | 3.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.5 | 4.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.5 | 1.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.5 | 2.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 3.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.5 | 2.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 2.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.5 | 2.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.5 | 5.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 1.4 | GO:0002159 | desmosome assembly(GO:0002159) response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.4 | 1.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.4 | 2.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.4 | 1.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.4 | 9.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.4 | 6.6 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.4 | 0.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.4 | 0.9 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.4 | 1.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 16.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.4 | 0.8 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.4 | 15.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.4 | 1.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.4 | 1.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 3.7 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.4 | 0.8 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.4 | 2.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.4 | 23.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.4 | 10.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.4 | 0.8 | GO:1901654 | response to ketone(GO:1901654) cellular response to ketone(GO:1901655) |
| 0.4 | 0.8 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.4 | 0.8 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.4 | 6.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 4.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.4 | 0.7 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.4 | 2.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.4 | 9.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.4 | 5.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.4 | 1.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.4 | 60.1 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.4 | 1.4 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) |
| 0.4 | 0.7 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.4 | 4.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 5.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 5.5 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.3 | 3.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.3 | 1.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.3 | 1.0 | GO:0035482 | gastric motility(GO:0035482) gastric emptying(GO:0035483) negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 2.9 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 2.3 | GO:0098885 | maternal process involved in parturition(GO:0060137) modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.3 | 2.3 | GO:0044838 | cell quiescence(GO:0044838) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 1.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.3 | 5.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 1.6 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.3 | 1.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 1.9 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 33.2 | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.3 | 0.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 5.9 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.3 | 3.7 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.3 | 2.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 1.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 2.6 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.3 | 2.9 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 2.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 10.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.3 | 3.1 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.3 | 2.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 0.8 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.3 | 0.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 1.6 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 1.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 0.8 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.3 | 3.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 5.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 0.8 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.3 | 0.8 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.3 | 1.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 8.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.3 | 1.5 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.3 | 3.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 1.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.2 | 1.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 3.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 4.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.5 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.2 | 1.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 0.2 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 1.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 0.7 | GO:0021830 | substrate-independent telencephalic tangential migration(GO:0021826) interneuron migration from the subpallium to the cortex(GO:0021830) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.2 | 0.7 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.2 | 2.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 3.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 0.9 | GO:0006586 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) |
| 0.2 | 3.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 1.7 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 5.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 1.0 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.2 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 1.5 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 1.5 | GO:1903044 | protein localization to membrane raft(GO:1903044) |
| 0.2 | 1.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 3.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 0.8 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.2 | 0.8 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.2 | 1.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 2.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.2 | 0.6 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.2 | 0.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 2.6 | GO:0023058 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.2 | 0.9 | GO:0097646 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 1.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.2 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.2 | 0.7 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 | 0.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 5.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.2 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 12.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 0.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 2.6 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.2 | 5.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.2 | 1.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 6.2 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.2 | 16.7 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.2 | 1.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 2.9 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 3.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 1.0 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 0.8 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 7.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.2 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.2 | 1.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 1.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 1.6 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.2 | 1.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 4.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 0.8 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 1.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 0.6 | GO:0035826 | rubidium ion transport(GO:0035826) cellular hypotonic response(GO:0071476) |
| 0.2 | 0.6 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 0.8 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.6 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.7 | GO:0036438 | regulation of heterotypic cell-cell adhesion(GO:0034114) positive regulation of heterotypic cell-cell adhesion(GO:0034116) maintenance of lens transparency(GO:0036438) |
| 0.1 | 3.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.1 | 1.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.5 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 3.8 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 9.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 1.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 6.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 3.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.6 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.1 | 0.4 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.5 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.2 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 1.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.6 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 8.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 2.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.7 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.5 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 1.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 1.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.5 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 1.1 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 2.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.8 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 1.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 4.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.3 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 2.2 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 0.3 | GO:2000778 | positive regulation of interleukin-6 secretion(GO:2000778) |
| 0.1 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.4 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 2.1 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.8 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 1.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 4.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 4.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 1.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 2.6 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.5 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.1 | 0.8 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.4 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 1.0 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 6.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 3.6 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 3.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 3.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.6 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.6 | GO:0033233 | regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.4 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 0.4 | GO:0044819 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.0 | 1.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.2 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 2.0 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.9 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0043512 | inhibin A complex(GO:0043512) |
| 4.6 | 13.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 4.4 | 17.8 | GO:0031673 | H zone(GO:0031673) |
| 3.5 | 17.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 3.2 | 29.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 2.9 | 26.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 2.7 | 51.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 2.2 | 44.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 2.2 | 32.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.9 | 21.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.9 | 78.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.8 | 5.4 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 1.8 | 26.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.7 | 8.5 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.7 | 22.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.7 | 33.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.5 | 5.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.4 | 28.9 | GO:0031430 | M band(GO:0031430) |
| 1.3 | 5.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.2 | 20.8 | GO:0031045 | dense core granule(GO:0031045) |
| 1.2 | 17.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.1 | 4.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.0 | 4.0 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.0 | 3.0 | GO:0098835 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 1.0 | 2.9 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.9 | 10.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.9 | 14.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.9 | 11.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.8 | 3.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 7.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.8 | 19.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 9.4 | GO:0031672 | A band(GO:0031672) |
| 0.7 | 14.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 8.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.7 | 159.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.6 | 3.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 1.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.6 | 58.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.5 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 4.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.5 | 12.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.5 | 10.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 5.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.5 | 1.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 4.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 3.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.4 | 14.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 4.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.4 | 51.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.4 | 19.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 41.3 | GO:0030017 | sarcomere(GO:0030017) |
| 0.3 | 2.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 4.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 1.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 2.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 2.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 1.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 9.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.3 | 6.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 26.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.3 | 2.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.3 | 13.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.3 | 1.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 9.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 1.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 20.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 1.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 7.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.2 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 1.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 4.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 2.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 4.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.2 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.9 | GO:1903440 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 3.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 1.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 4.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 2.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 2.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 36.4 | GO:0098794 | postsynapse(GO:0098794) |
| 0.2 | 8.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 2.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 101.9 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 10.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 6.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 3.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 9.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 13.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 4.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 9.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 7.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 10.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 5.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.7 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 25.6 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 1.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 5.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 15.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 4.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.9 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.1 | 3.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 9.1 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 3.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 11.1 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 1.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 5.7 | 34.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 5.3 | 21.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.3 | 17.4 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 3.8 | 30.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 3.4 | 17.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 3.2 | 25.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 3.2 | 9.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 2.9 | 11.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 2.6 | 10.5 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 2.3 | 36.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 2.2 | 8.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.2 | 22.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 2.2 | 8.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 2.1 | 10.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 2.1 | 12.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.9 | 7.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 1.9 | 5.6 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.9 | 11.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.8 | 60.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 1.8 | 7.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 1.7 | 14.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.6 | 1.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 1.6 | 12.6 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 1.5 | 87.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 1.4 | 7.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 1.4 | 17.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.4 | 33.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.4 | 41.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.4 | 16.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.3 | 33.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 1.3 | 13.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.3 | 4.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 1.3 | 5.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.3 | 2.6 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.3 | 3.8 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.2 | 71.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.2 | 8.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.2 | 3.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.2 | 5.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.1 | 12.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 1.1 | 19.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.0 | 3.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.0 | 3.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.0 | 3.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.0 | 3.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.0 | 30.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.9 | 28.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.9 | 3.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.9 | 6.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.9 | 16.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.9 | 6.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.9 | 3.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.9 | 15.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.8 | 0.8 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.8 | 5.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 2.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.8 | 4.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.8 | 11.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.8 | 3.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.8 | 3.8 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.8 | 2.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.8 | 9.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.7 | 8.1 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.7 | 12.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 2.8 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.7 | 3.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.7 | 11.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.7 | 2.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.6 | 14.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.6 | 4.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.6 | 10.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.6 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.6 | 2.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 4.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 2.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.6 | 7.0 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.6 | 1.7 | GO:0019966 | C-X-C chemokine binding(GO:0019958) interleukin-1 binding(GO:0019966) tumor necrosis factor binding(GO:0043120) |
| 0.6 | 1.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.6 | 3.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 2.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.6 | 12.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.5 | 12.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 2.6 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 26.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.5 | 4.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 2.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 12.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.5 | 1.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.5 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 1.4 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.4 | 1.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.4 | 4.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 2.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 2.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.4 | 4.9 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.4 | 3.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 3.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 1.8 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.4 | 2.6 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.4 | 1.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.4 | 23.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.3 | 1.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 2.4 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.3 | 1.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.3 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 6.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.3 | 2.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 0.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 4.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.3 | 56.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.3 | 2.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.3 | 14.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.3 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 0.9 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 2.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.3 | 0.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 2.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 2.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 3.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.9 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 29.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.3 | 1.3 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.3 | 10.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 7.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 2.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 1.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 3.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 12.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 58.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 5.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 3.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 1.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 2.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 5.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 3.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 15.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 2.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 3.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 8.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 1.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 20.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 4.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.2 | 0.9 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 1.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 1.8 | GO:0042562 | hormone binding(GO:0042562) |
| 0.2 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 4.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 1.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 3.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 0.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 15.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.2 | 1.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 4.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 2.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 5.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 3.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 5.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 4.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.4 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.8 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 2.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 2.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 27.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.8 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 3.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 11.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.5 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 25.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 1.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 12.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 1.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 2.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 2.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 3.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.5 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 2.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 9.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 2.2 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 81.2 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 1.0 | 43.9 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.9 | 31.1 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.8 | 9.3 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.8 | 29.9 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.6 | 3.6 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.6 | 8.3 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.5 | 17.9 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.5 | 7.1 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.5 | 19.9 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.5 | 19.4 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 5.1 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.5 | 16.7 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.4 | 12.7 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.4 | 18.9 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 9.3 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 24.3 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.4 | 21.8 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.3 | 15.1 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 9.5 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 14.9 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.3 | 14.9 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 11.1 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.3 | 10.8 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.3 | 6.2 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 7.9 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.2 | 4.5 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 3.5 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.2 | 1.5 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 4.7 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.2 | 3.0 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 16.4 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 6.2 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.2 | 6.3 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.2 | 2.5 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.2 | 3.2 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 6.0 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.2 | 1.2 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 8.0 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.5 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.2 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.7 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 5.0 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 8.6 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 15.1 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.9 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.2 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.9 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.9 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.9 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.2 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 1.1 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.6 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.3 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.1 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 0.2 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.2 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.8 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.2 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 2.5 | 25.4 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 2.0 | 71.9 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 2.0 | 84.0 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 1.7 | 46.6 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 1.5 | 18.9 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 1.4 | 37.6 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 1.2 | 22.9 | REACTOME_LIGAND_GATED_ION_CHANNEL_TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.8 | 21.4 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.7 | 9.0 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.7 | 12.9 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.7 | 4.1 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 8.4 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.6 | 7.7 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 6.7 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.6 | 16.0 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.6 | 8.1 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.6 | 45.3 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.6 | 24.6 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.5 | 3.1 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 3.5 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.4 | 1.3 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.4 | 3.9 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 7.7 | REACTOME_GRB2_EVENTS_IN_ERBB2_SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.4 | 7.7 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.4 | 3.2 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.4 | 35.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 8.1 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.3 | 6.7 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 7.3 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 10.6 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 21.3 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 2.2 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 2.4 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 1.3 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 2.2 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.2 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 2.9 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 2.0 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 1.5 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 7.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 1.7 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 1.6 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 2.3 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 3.2 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 1.5 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 2.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.1 | REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 3.2 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 3.1 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.5 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.7 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.5 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 11.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.9 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 6.2 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.9 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.9 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 6.2 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.4 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.5 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 5.0 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.8 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.5 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.4 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.1 | REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 3.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.7 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.8 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.4 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.1 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.2 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 1.3 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 0.9 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.5 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.5 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.8 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.5 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.8 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.5 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME_G1_PHASE | Genes involved in G1 Phase |