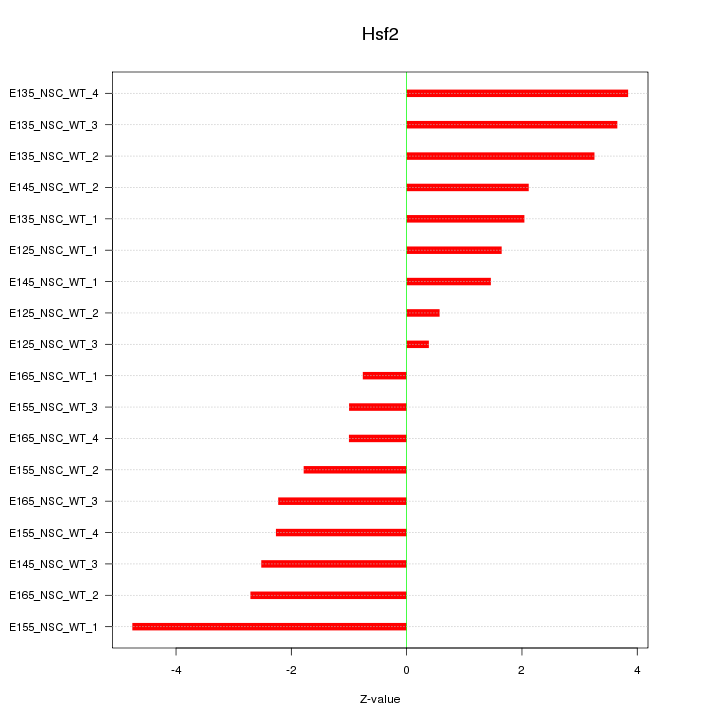
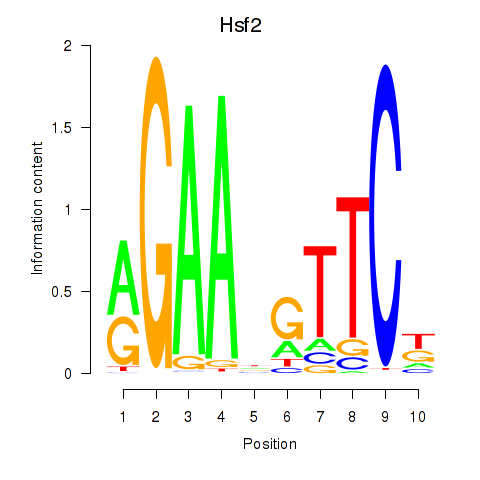
Motif ID: Hsf2
Z-value: 2.414
Transcription factors associated with Hsf2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hsf2 | ENSMUSG00000019878.7 | Hsf2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | mm10_v2_chr10_+_57486354_57486414 | 0.03 | 9.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:2000981 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 1.3 | 5.4 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.2 | 3.7 | GO:0030421 | defecation(GO:0030421) |
| 1.2 | 4.7 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 1.0 | 4.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 1.0 | 3.0 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 1.0 | 3.9 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.0 | 3.8 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.9 | 15.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.9 | 3.7 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) positive regulation of NK T cell activation(GO:0051135) |
| 0.9 | 3.6 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.9 | 2.6 | GO:0003360 | brainstem development(GO:0003360) |
| 0.8 | 2.5 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.8 | 3.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.8 | 2.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.7 | 2.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.7 | 4.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 4.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.7 | 3.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.6 | 1.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.6 | 2.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.6 | 3.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.6 | 1.9 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.6 | 1.8 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.6 | 1.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.5 | 1.6 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.5 | 1.5 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.5 | 3.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 3.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.5 | 1.9 | GO:0060032 | notochord regression(GO:0060032) |
| 0.5 | 3.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.5 | 1.4 | GO:1903054 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) negative regulation of extracellular matrix organization(GO:1903054) positive regulation of protein folding(GO:1903334) |
| 0.5 | 4.1 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 | 1.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 2.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.4 | 1.7 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.4 | 1.3 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.4 | 1.3 | GO:0097278 | transforming growth factor beta activation(GO:0036363) complement-dependent cytotoxicity(GO:0097278) |
| 0.4 | 2.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.4 | 2.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.4 | 1.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.4 | 1.6 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.4 | 3.6 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.4 | 3.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.4 | 1.8 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.4 | 2.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.4 | 1.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.4 | 3.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.4 | 1.4 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.4 | 1.1 | GO:1904154 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 0.7 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 | 1.3 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.3 | 2.8 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 0.9 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.3 | 0.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 2.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 0.8 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.3 | 1.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 0.8 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.3 | 1.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 0.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 1.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 1.6 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 | 1.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 1.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.3 | 0.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.3 | 0.8 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.3 | 2.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 3.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 2.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 1.2 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 6.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 2.6 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 0.9 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 | 2.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 3.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 1.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 4.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 0.6 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.2 | 1.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 1.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 2.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 0.6 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 1.2 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 1.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 0.4 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.2 | 2.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.2 | 0.6 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 | 0.4 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) |
| 0.2 | 0.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 0.9 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.2 | 0.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 1.4 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.2 | 0.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 2.0 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 | 1.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 0.5 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.2 | 0.8 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.2 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 5.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 9.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 1.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 1.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 0.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 0.8 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 3.9 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 5.4 | GO:1902750 | negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 | 0.3 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 1.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 2.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.4 | GO:0097026 | myeloid dendritic cell chemotaxis(GO:0002408) dendritic cell dendrite assembly(GO:0097026) mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 2.1 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 2.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 1.5 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 | 0.4 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.5 | GO:0061450 | trophoblast cell migration(GO:0061450) regulation of trophoblast cell migration(GO:1901163) negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.8 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 1.4 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.5 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 4.2 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 | 3.3 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.1 | 1.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.8 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.2 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.1 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.6 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 0.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:1904706 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 1.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.6 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 0.2 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.5 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 | 1.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 3.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 1.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.5 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.8 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 0.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 1.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.9 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 1.4 | GO:0048636 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) |
| 0.0 | 0.7 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.2 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 0.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.4 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.3 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 1.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 1.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.6 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 1.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 2.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 3.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.4 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.4 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.0 | 1.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.9 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.6 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.2 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.0 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.0 | 0.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.8 | 3.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.7 | 4.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.6 | 2.9 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.6 | 3.4 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.5 | 3.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.5 | 1.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.5 | 2.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 3.9 | GO:0005818 | aster(GO:0005818) |
| 0.5 | 2.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 2.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 1.6 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.4 | 1.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 5.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 1.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 3.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 2.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 1.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 1.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 0.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 0.6 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 1.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 2.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 8.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.1 | 1.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.9 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 8.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 5.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.6 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 2.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 4.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 4.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.2 | GO:0071014 | U2-type post-mRNA release spliceosomal complex(GO:0071008) post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.7 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 3.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 5.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 3.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 2.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.8 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.8 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 8.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.4 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 6.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 5.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 16.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.1 | 3.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.9 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.8 | 3.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.7 | 15.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.7 | 2.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.6 | 1.9 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.6 | 3.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.5 | 2.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.5 | 2.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.5 | 2.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 3.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 2.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.5 | 1.4 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.4 | 1.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.4 | 3.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.4 | 4.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 2.6 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.4 | 2.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.4 | 1.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.4 | 3.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 3.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 1.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 1.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 5.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 1.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 2.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 3.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.3 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.3 | 1.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 1.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 0.9 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.3 | 2.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 0.8 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 0.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 1.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 1.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.3 | 2.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 1.8 | GO:0031419 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) cobalamin binding(GO:0031419) |
| 0.3 | 4.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 3.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.7 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.2 | 1.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 1.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.2 | 1.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 3.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 1.6 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.2 | 6.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 3.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 3.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 1.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 1.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 5.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 0.5 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.2 | 1.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 4.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 1.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.9 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 2.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 4.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 5.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 3.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.9 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0052740 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 4.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 1.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.0 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 2.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 1.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 5.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 1.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 3.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 0.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 5.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 2.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 2.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 1.0 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.4 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.8 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 4.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.1 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 6.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.7 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 2.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 23.9 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 6.6 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.2 | 6.8 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 6.3 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.2 | 5.9 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 2.5 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.2 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.4 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 4.2 | PID_IFNG_PATHWAY | IFN-gamma pathway |
| 0.1 | 5.3 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.1 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 8.6 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.4 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 1.6 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 1.9 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.1 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 10.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.1 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 1.6 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.7 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.1 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 1.0 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.1 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.7 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.9 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.1 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.3 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.2 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 3.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.9 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.2 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 3.6 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 4.1 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 2.5 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 3.0 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.3 | 0.5 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 7.8 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.4 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 3.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.1 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 1.3 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 1.4 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 1.9 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 3.6 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.2 | 3.4 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 6.5 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 1.8 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 5.7 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.8 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 5.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 2.7 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 7.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.4 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 6.6 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.9 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.4 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.5 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 2.8 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.8 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.0 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 2.1 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.3 | REACTOME_INTERFERON_GAMMA_SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.4 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.9 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.3 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.4 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.9 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.2 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.2 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME_P53_DEPENDENT_G1_DNA_DAMAGE_RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.9 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 1.9 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 5.0 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.1 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME_TRANSPORT_OF_GLUCOSE_AND_OTHER_SUGARS_BILE_SALTS_AND_ORGANIC_ACIDS_METAL_IONS_AND_AMINE_COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 1.5 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME_TRNA_AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.3 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.1 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.1 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.3 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.2 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.9 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |