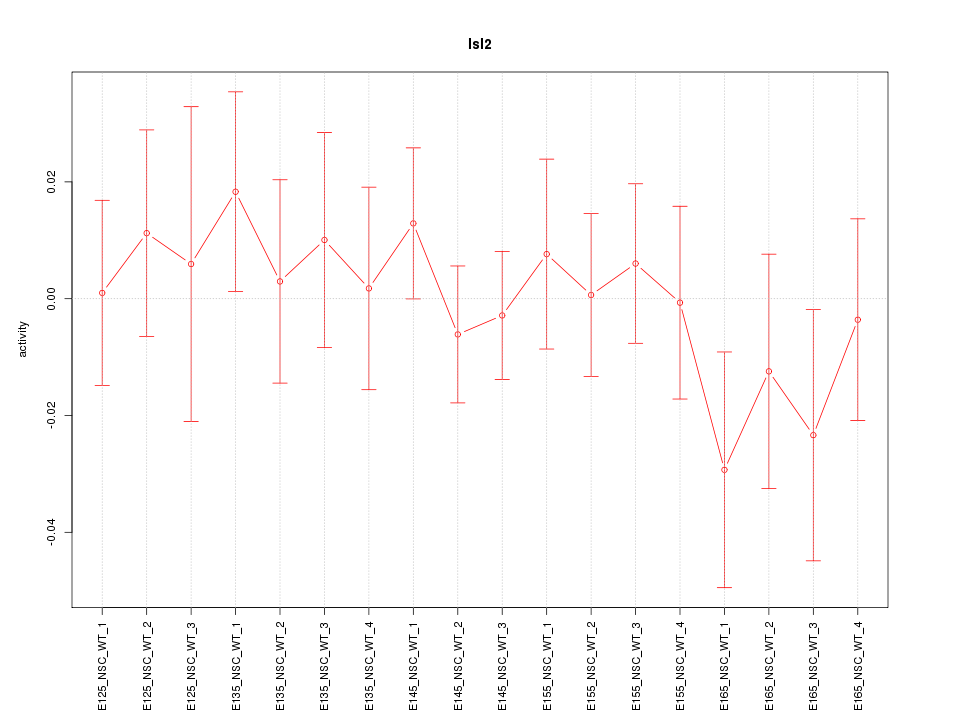
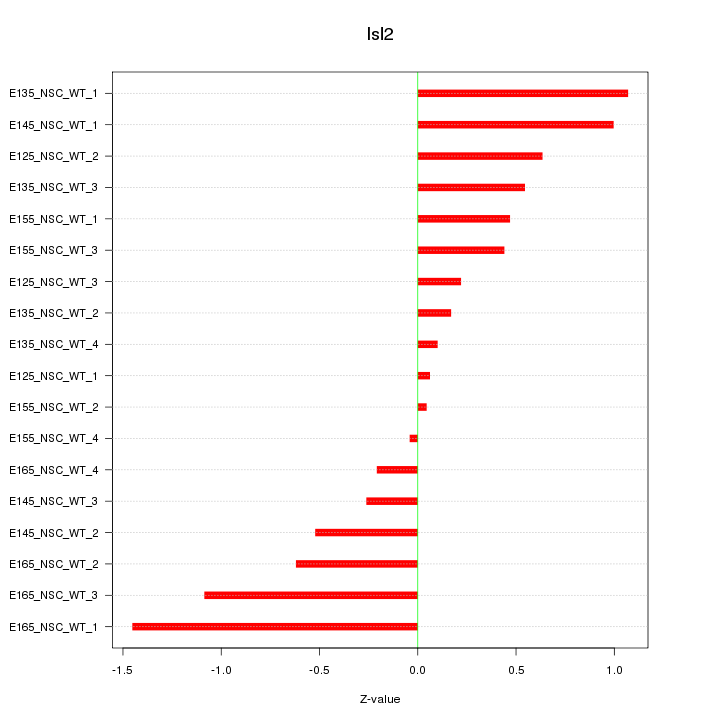
Motif ID: Isl2
Z-value: 0.641
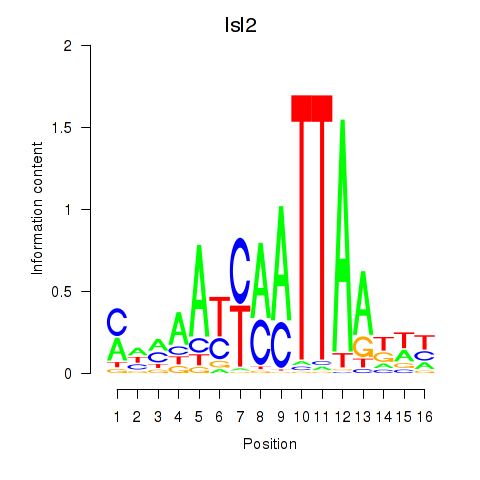
Transcription factors associated with Isl2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Isl2 | ENSMUSG00000032318.6 | Isl2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 2.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.5 | 1.5 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.4 | 1.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 1.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.7 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 0.6 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 0.4 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 0.7 | GO:0072592 | regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.4 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.8 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.6 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 1.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 1.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.3 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.4 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 0.3 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.3 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 1.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0045835 | meiotic cell cycle checkpoint(GO:0033313) negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 0.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.2 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 2.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 1.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.4 | 2.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 1.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 1.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.9 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.8 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 2.1 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID_FGF_PATHWAY | FGF signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 0.8 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.5 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.5 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.7 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.2 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |