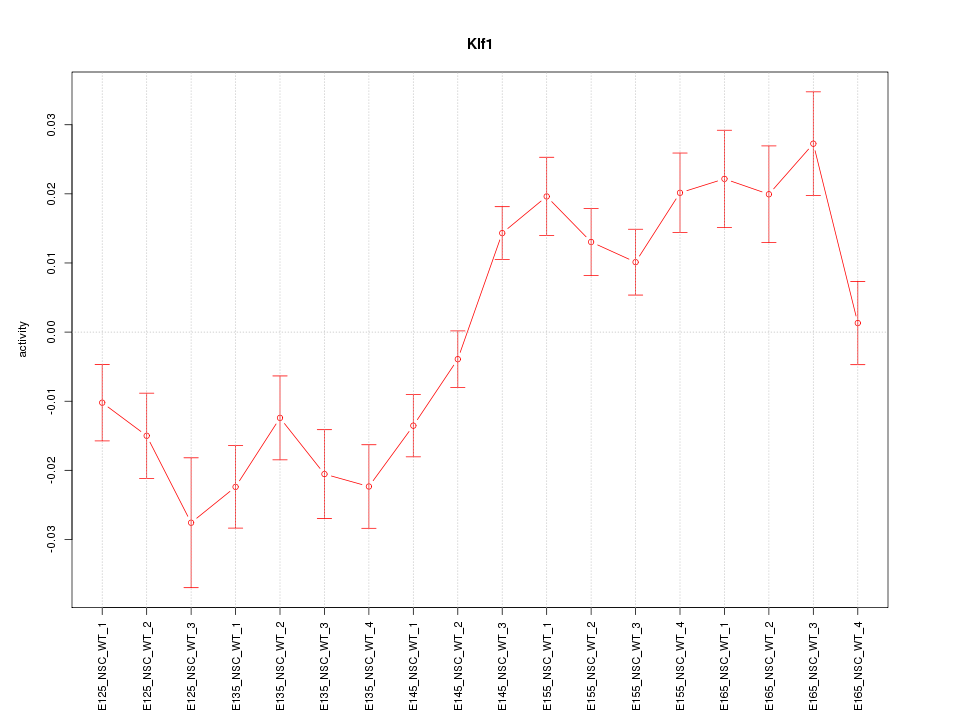
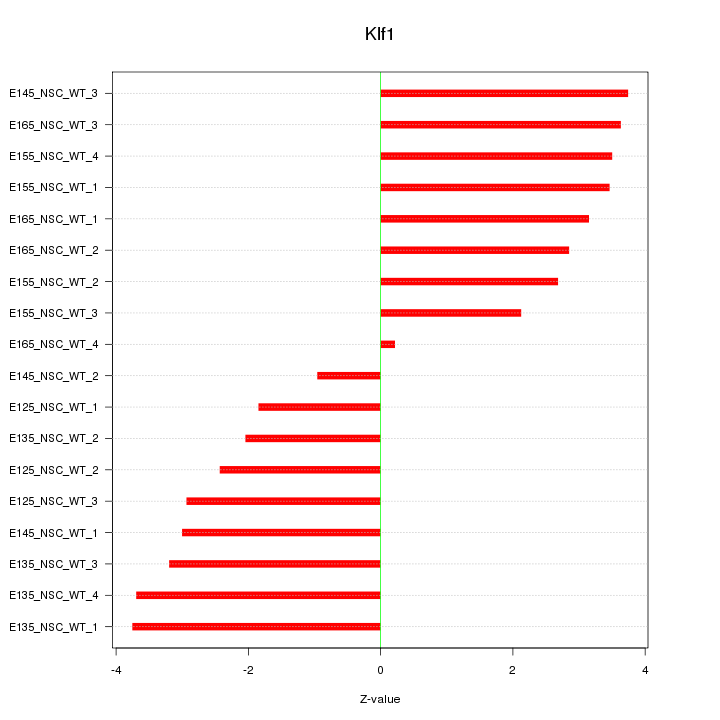
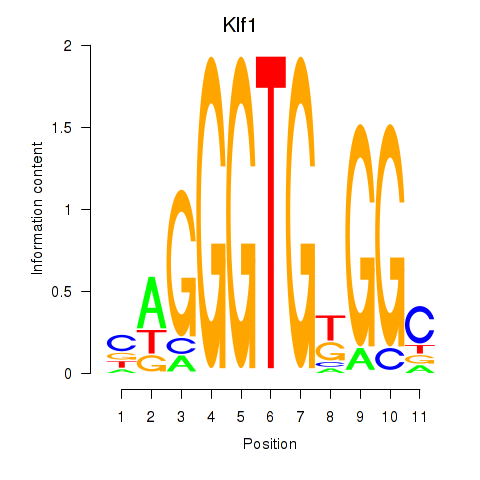
Motif ID: Klf1
Z-value: 2.899
Transcription factors associated with Klf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Klf1 | ENSMUSG00000054191.7 | Klf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf1 | mm10_v2_chr8_+_84901928_84901992 | -0.79 | 8.4e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 3.3 | 9.8 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 3.0 | 27.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 2.4 | 9.5 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 2.1 | 6.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 2.0 | 10.2 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.8 | 9.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 1.6 | 14.8 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 1.4 | 8.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 1.4 | 6.8 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.4 | 13.5 | GO:0046959 | habituation(GO:0046959) |
| 1.2 | 3.6 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.2 | 1.2 | GO:0072197 | renal vesicle induction(GO:0072034) ureter morphogenesis(GO:0072197) |
| 1.2 | 11.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 1.2 | 5.8 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 1.1 | 1.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 1.1 | 3.4 | GO:1904978 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 1.1 | 6.7 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 1.1 | 1.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 | 3.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.0 | 3.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.0 | 1.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 1.0 | 5.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.0 | 2.9 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 1.0 | 5.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 1.0 | 2.9 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.0 | 1.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 1.0 | 2.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.0 | 5.7 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.9 | 0.9 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.9 | 4.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.9 | 0.9 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.8 | 7.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.8 | 4.2 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.8 | 2.4 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.8 | 4.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.8 | 3.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.8 | 2.4 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.8 | 2.3 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.7 | 8.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.7 | 2.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.7 | 2.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.7 | 2.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.7 | 2.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.7 | 2.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.7 | 5.4 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.7 | 2.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.7 | 2.0 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.7 | 5.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.7 | 2.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.6 | 1.9 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.6 | 3.9 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.6 | 1.9 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.6 | 3.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.6 | 3.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.6 | 1.9 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.6 | 1.9 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.6 | 2.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.6 | 2.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.6 | 1.8 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.6 | 2.4 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.6 | 0.6 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.6 | 4.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.6 | 1.7 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.6 | 4.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.6 | 10.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.6 | 1.7 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.6 | 6.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.6 | 1.7 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.5 | 1.6 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.5 | 1.6 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.5 | 2.7 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.5 | 1.6 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.5 | 1.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.5 | 3.7 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.5 | 2.6 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.5 | 5.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.5 | 1.5 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.5 | 9.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.5 | 2.6 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.5 | 4.1 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.5 | 2.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.5 | 6.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.5 | 0.5 | GO:1901608 | regulation of vesicle transport along microtubule(GO:1901608) |
| 0.5 | 2.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.5 | 2.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.5 | 5.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.5 | 2.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 1.5 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.5 | 0.5 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.5 | 7.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.5 | 2.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.5 | 1.9 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.5 | 5.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 0.9 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.5 | 2.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.5 | 3.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 1.4 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.5 | 1.4 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.5 | 3.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.5 | 1.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.5 | 1.4 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.5 | 0.9 | GO:0046878 | regulation of saliva secretion(GO:0046877) positive regulation of saliva secretion(GO:0046878) |
| 0.5 | 1.8 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.5 | 3.2 | GO:0035826 | rubidium ion transport(GO:0035826) |
| 0.4 | 3.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.4 | 4.0 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.4 | 1.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 2.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.4 | 2.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 2.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 3.9 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.4 | 0.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 0.9 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.4 | 1.3 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.4 | 8.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 1.3 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.4 | 2.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.4 | 2.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.4 | 1.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 4.9 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.4 | 1.2 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.4 | 6.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.4 | 1.6 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.4 | 1.2 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.4 | 0.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 3.5 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.4 | 2.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 3.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.4 | 3.5 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.4 | 1.5 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.4 | 2.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.4 | 2.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.4 | 1.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 1.5 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.4 | 3.0 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.4 | 1.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.4 | 1.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.4 | 1.5 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 | 4.0 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.4 | 0.4 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.4 | 2.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.4 | 2.9 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.4 | 8.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.4 | 2.5 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.4 | 1.4 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.4 | 1.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.4 | 1.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 0.4 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.4 | 0.7 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.4 | 1.1 | GO:0072720 | cellular response to UV-A(GO:0071492) response to dithiothreitol(GO:0072720) |
| 0.4 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 0.7 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.4 | 2.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.4 | 3.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 2.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 1.4 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.3 | 1.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.3 | 1.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 7.0 | GO:0048265 | response to pain(GO:0048265) |
| 0.3 | 7.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 9.1 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.3 | 0.6 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.3 | 2.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 1.3 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 0.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 1.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 4.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 2.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 2.2 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.3 | 1.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.3 | 1.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 0.6 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 0.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.2 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.3 | 1.2 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.3 | 2.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 6.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 0.9 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 1.2 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.3 | 1.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.3 | 1.7 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.3 | 3.7 | GO:0043482 | pigment accumulation(GO:0043476) cellular pigment accumulation(GO:0043482) |
| 0.3 | 0.9 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.3 | 4.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 4.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 3.3 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.3 | 1.6 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 0.8 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.3 | 0.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 1.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 1.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.3 | 0.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 2.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.3 | 1.8 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 | 0.8 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 | 3.3 | GO:0072319 | vesicle uncoating(GO:0072319) |
| 0.3 | 1.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 | 1.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 0.7 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 1.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.5 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.2 | 1.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 1.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 0.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 1.4 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.2 | 0.9 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.2 | 1.6 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.2 | 2.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 1.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 1.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 0.5 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 1.1 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.2 | 1.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 0.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 4.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.7 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 4.9 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 0.7 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 0.7 | GO:1900365 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 2.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 1.5 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.2 | 0.9 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 2.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 0.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.2 | 1.7 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 3.5 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.2 | 0.6 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 0.4 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.2 | 0.9 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 3.0 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.2 | 0.4 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 0.2 | 0.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 0.6 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.2 | 1.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.2 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 4.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 5.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.2 | 2.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.2 | 1.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.2 | 0.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 0.2 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.2 | 1.2 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.2 | 1.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 0.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 2.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 0.6 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 0.4 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.2 | 1.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.8 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 4.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 14.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 1.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.2 | 0.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 1.5 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.2 | 0.4 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
| 0.2 | 1.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.2 | 1.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.2 | 0.2 | GO:0072610 | interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 0.6 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.2 | 0.6 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.2 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 | 0.4 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.7 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 0.6 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.7 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.2 | 1.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 0.5 | GO:0070255 | mucus secretion(GO:0070254) regulation of mucus secretion(GO:0070255) |
| 0.2 | 4.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 0.7 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 0.5 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.2 | 0.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 0.5 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 4.8 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 1.4 | GO:2000489 | hepatic stellate cell activation(GO:0035733) regulation of hepatic stellate cell activation(GO:2000489) |
| 0.2 | 0.7 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.2 | 1.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 5.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 2.9 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 1.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 0.5 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.2 | 4.6 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.2 | 2.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 2.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 2.0 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.2 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.5 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 | 2.3 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.2 | 1.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 5.7 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.2 | 1.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.8 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 2.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 1.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.8 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.2 | 0.6 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 0.8 | GO:0071635 | negative regulation of transforming growth factor beta1 production(GO:0032911) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.2 | 1.7 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 2.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.4 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.2 | 0.6 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.2 | 4.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 1.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.6 | GO:0040010 | B-1 B cell homeostasis(GO:0001922) growth involved in heart morphogenesis(GO:0003241) positive regulation of growth rate(GO:0040010) |
| 0.2 | 1.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.5 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.2 | 2.7 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 0.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 2.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 1.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.6 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 2.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 10.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.4 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.1 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.8 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.5 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 1.5 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 2.0 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 0.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 1.1 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.4 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 0.5 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.0 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 0.6 | GO:0055119 | relaxation of cardiac muscle(GO:0055119) |
| 0.1 | 0.3 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.1 | 1.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 1.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 3.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.1 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 1.0 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.7 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 1.2 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.2 | GO:1902256 | endocardial cushion fusion(GO:0003274) apoptotic process involved in outflow tract morphogenesis(GO:0003275) atrial septum primum morphogenesis(GO:0003289) negative regulation of macrophage cytokine production(GO:0010936) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) positive regulation of GTP binding(GO:1904426) positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905007) |
| 0.1 | 0.2 | GO:1901142 | insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.1 | 0.6 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 1.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.1 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 1.5 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 2.9 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 3.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 4.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 1.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 1.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.5 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.7 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.3 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.2 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 1.0 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 1.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 2.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.4 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.1 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 0.1 | 0.7 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 1.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.7 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.3 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 4.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.8 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 0.4 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.3 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) sequestering of triglyceride(GO:0030730) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.3 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.3 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0046543 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.5 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.4 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 2.2 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.1 | 1.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.1 | GO:0070142 | synaptic vesicle budding(GO:0070142) |
| 0.1 | 1.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.1 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.3 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:0050904 | diapedesis(GO:0050904) |
| 0.1 | 0.6 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.9 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.1 | 7.4 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 7.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.0 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.1 | 1.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 3.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 2.0 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.1 | 2.3 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.1 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.7 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 1.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.7 | GO:0061097 | regulation of protein tyrosine kinase activity(GO:0061097) |
| 0.1 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.3 | GO:1903028 | asymmetric Golgi ribbon formation(GO:0090164) regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.9 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.5 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.1 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.1 | 0.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 1.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 2.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.2 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.4 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.2 | GO:0019935 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 | 0.8 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.0 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.3 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.0 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.4 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.2 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.1 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0031652 | positive regulation of heat generation(GO:0031652) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.3 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 1.1 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.4 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 2.1 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 2.0 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0010453 | regulation of cell fate commitment(GO:0010453) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 1.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.3 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0006272 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.0 | 0.2 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 1.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0001941 | postsynaptic membrane organization(GO:0001941) |
| 0.0 | 0.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 2.8 | 11.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 2.7 | 10.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 2.4 | 23.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.0 | 9.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.4 | 5.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.3 | 1.3 | GO:0019034 | viral replication complex(GO:0019034) |
| 1.1 | 4.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.0 | 3.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.0 | 2.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.9 | 3.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.8 | 5.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 8.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.7 | 2.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.7 | 12.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 11.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 2.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.7 | 4.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.6 | 5.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.6 | 2.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.6 | 7.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.6 | 1.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.5 | 1.6 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.5 | 1.6 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.5 | 4.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 1.5 | GO:0071920 | cleavage body(GO:0071920) |
| 0.5 | 1.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.5 | 6.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 1.9 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.5 | 4.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 24.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.4 | 5.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 1.3 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.4 | 4.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 3.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 12.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.4 | 1.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.4 | 6.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.4 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 0.8 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.4 | 1.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.4 | 4.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.4 | 1.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.4 | 3.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 2.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 6.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 28.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.3 | 3.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 3.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 0.3 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 2.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 2.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 4.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 3.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 4.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 1.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 1.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 1.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 0.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 4.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 1.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 8.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 1.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 1.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.2 | 3.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 1.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 5.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 2.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 1.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 1.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 5.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 0.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 1.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 6.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 3.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 6.5 | GO:0031231 | intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 1.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 9.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.6 | GO:0000799 | nuclear condensin complex(GO:0000799) germinal vesicle(GO:0042585) |
| 0.2 | 1.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 0.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 2.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 0.6 | GO:0005940 | septin ring(GO:0005940) |
| 0.2 | 1.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.2 | 1.5 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.2 | 0.7 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 16.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 9.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.9 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 9.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 1.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 0.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 1.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 6.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 3.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 7.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 5.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 31.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 2.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 5.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 2.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 3.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.9 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 2.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 17.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 2.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 7.5 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 5.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 6.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 2.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 10.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 1.0 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 0.5 | GO:0044754 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.1 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 8.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 1.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 8.7 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 5.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.9 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 1.0 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 1.0 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 6.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 3.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 29.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 3.3 | 13.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 2.8 | 11.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 2.5 | 7.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.6 | 6.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.4 | 4.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.4 | 5.6 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 1.4 | 1.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 1.2 | 5.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 1.1 | 3.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 1.0 | 13.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.9 | 3.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.9 | 5.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.9 | 6.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.9 | 2.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.8 | 3.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.8 | 2.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.8 | 2.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.8 | 10.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.8 | 0.8 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.8 | 11.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.8 | 3.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.7 | 3.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.7 | 1.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.7 | 5.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 2.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.7 | 3.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.7 | 4.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 2.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.6 | 7.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.6 | 3.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.6 | 3.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 5.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.6 | 1.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 3.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.6 | 1.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.6 | 2.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 2.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.6 | 7.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.6 | 2.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.6 | 6.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 1.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.5 | 7.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 2.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 9.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.5 | 3.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.5 | 1.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.5 | 1.5 | GO:0031896 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) V2 vasopressin receptor binding(GO:0031896) |
| 0.5 | 1.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 7.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.5 | 6.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.5 | 4.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.5 | 9.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 3.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.5 | 0.5 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.5 | 2.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.5 | 1.9 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 1.4 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.5 | 0.9 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.5 | 8.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.4 | 1.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.4 | 5.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 2.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 7.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.4 | 1.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 2.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 1.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.4 | 2.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.4 | 7.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 1.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 2.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 4.0 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.4 | 1.6 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.4 | 1.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.4 | 1.9 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.4 | 4.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 3.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 1.1 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.4 | 1.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 9.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 3.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.4 | 1.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.4 | 1.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 1.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 1.4 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.4 | 2.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 2.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 1.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 3.4 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.3 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 3.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 4.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 1.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 3.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.9 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.3 | 0.9 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.3 | 0.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 4.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 2.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 2.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 20.8 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.3 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 1.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 0.9 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.3 | 1.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 1.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 2.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 10.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 1.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 2.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 3.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 0.7 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.2 | 13.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 3.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.5 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 0.7 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.2 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 2.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.9 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 0.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 0.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 4.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 7.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 5.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 4.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 0.6 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.2 | 0.6 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 0.6 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.2 | 0.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 1.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 4.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 1.0 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.2 | 0.8 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 2.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 2.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 4.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 3.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 3.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.9 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.2 | 0.5 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.7 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 4.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 8.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 1.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 6.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 1.0 | GO:0034595 | phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.2 | 2.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.2 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.2 | 2.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.2 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.9 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 2.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.6 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 5.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 9.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 11.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 4.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 37.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.4 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.4 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 2.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 5.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 3.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.6 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 4.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 2.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.3 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 10.2 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 3.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.1 | 3.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 3.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 4.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 4.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 2.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 3.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 4.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 2.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 2.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 3.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 3.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 11.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 5.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.6 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0050997 | quaternary ammonium group binding(GO:0050997) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0015932 | nucleobase-containing compound transmembrane transporter activity(GO:0015932) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.3 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 7.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 26.1 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 3.4 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.5 | 7.9 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.3 | 2.3 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.3 | 12.0 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.3 | 8.8 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.3 | 2.0 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 2.5 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 9.4 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 11.7 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.2 | 2.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.2 | 2.5 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.2 | 2.9 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 6.9 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 4.3 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.2 | 2.5 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.2 | 4.6 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 2.1 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 2.6 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 0.9 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 3.0 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 3.0 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.7 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.7 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 16.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.4 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.1 | 3.7 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 2.1 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.1 | 8.3 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 0.4 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 0.7 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 7.2 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 4.1 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.2 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.8 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.8 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.1 | 0.6 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.8 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.1 | 2.9 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.1 | 2.7 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.4 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.1 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.5 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 3.5 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.3 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 1.5 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.2 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 0.4 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.7 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.8 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.6 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.1 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.4 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | PID_BCR_5PATHWAY | BCR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 1.3 | 29.6 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.0 | 2.0 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.8 | 2.4 | REACTOME_SEROTONIN_RECEPTORS | Genes involved in Serotonin receptors |
| 0.7 | 3.7 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.6 | 4.5 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.6 | 6.0 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 24.0 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 5.2 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.5 | 12.5 | REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.5 | 19.4 | REACTOME_INWARDLY_RECTIFYING_K_CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.4 | 2.9 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 1.6 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 3.8 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.4 | 8.6 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 6.6 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 9.5 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.3 | 9.3 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 4.8 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 7.7 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 0.6 | REACTOME_DOWNSTREAM_SIGNALING_OF_ACTIVATED_FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.3 | 3.6 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.3 | 3.0 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 10.7 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 3.6 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 3.4 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 4.1 | REACTOME_SIGNALING_BY_NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.3 | 1.5 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 7.5 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 0.2 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 5.0 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 1.6 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 10.6 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 2.5 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 1.9 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 4.9 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 5.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 0.8 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 0.2 | REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 1.0 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 6.2 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 3.1 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 15.2 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 4.3 | REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 3.5 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 12.4 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 4.4 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 0.7 | REACTOME_IL_2_SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.2 | 1.2 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 1.1 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.2 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 2.1 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.6 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.9 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 3.2 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.1 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.7 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.2 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.6 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.3 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.1 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 11.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.8 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.8 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.8 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.6 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.1 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.8 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.1 | REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 1.0 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.7 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.2 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.7 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.5 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.4 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 6.4 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 0.9 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.0 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.1 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.1 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.7 | REACTOME_POST_NMDA_RECEPTOR_ACTIVATION_EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 1.1 | REACTOME_SIGNALLING_TO_RAS | Genes involved in Signalling to RAS |
| 0.1 | 1.7 | REACTOME_PLC_BETA_MEDIATED_EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 0.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.5 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.6 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.3 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.9 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 0.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.8 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 0.6 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.6 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.1 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.7 | REACTOME_COMPLEMENT_CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.7 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.7 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.2 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.6 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME_BOTULINUM_NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.1 | REACTOME_SIGNALING_BY_ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |