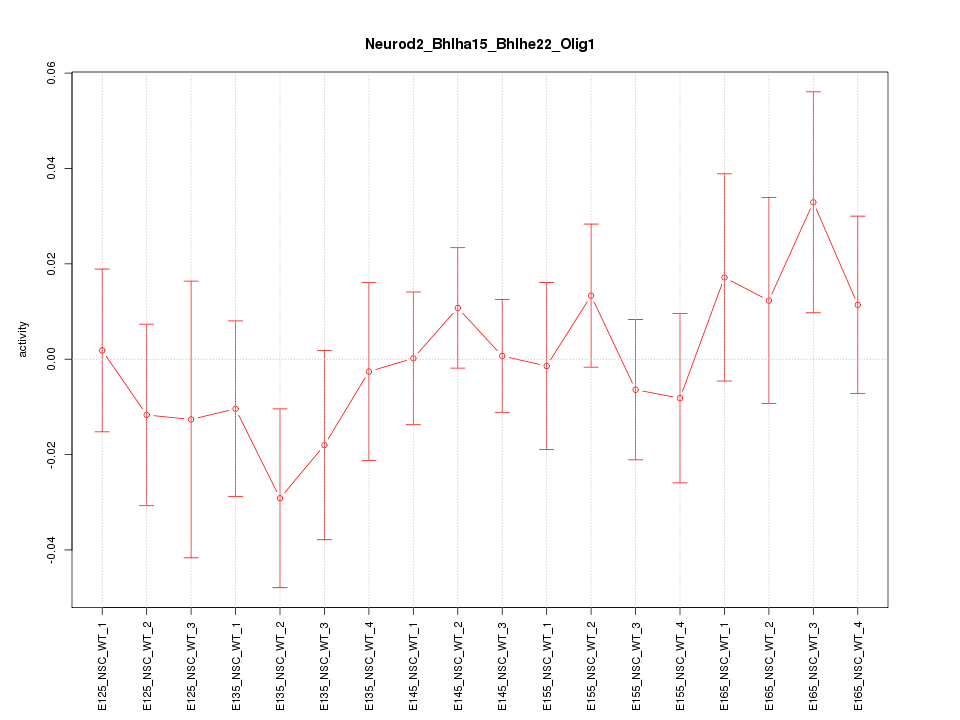
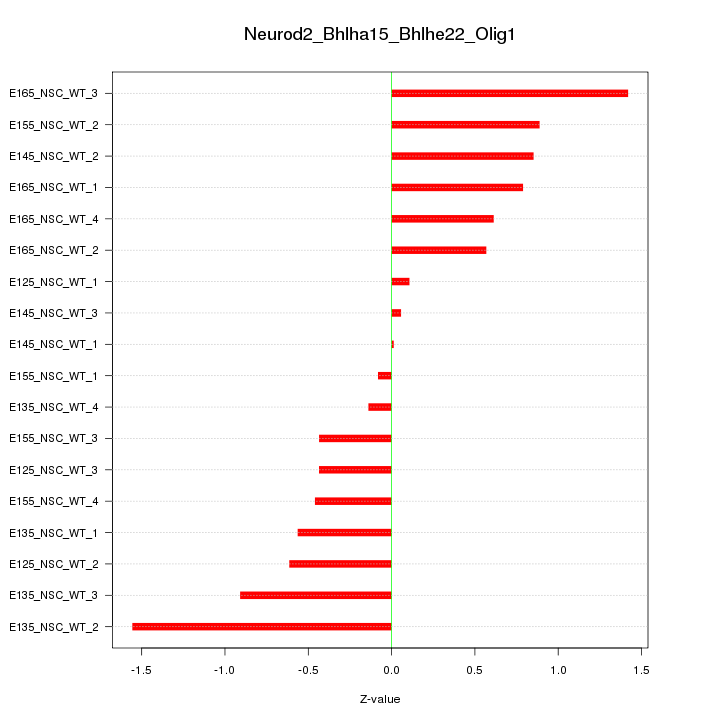
Motif ID: Neurod2_Bhlha15_Bhlhe22_Olig1
Z-value: 0.724
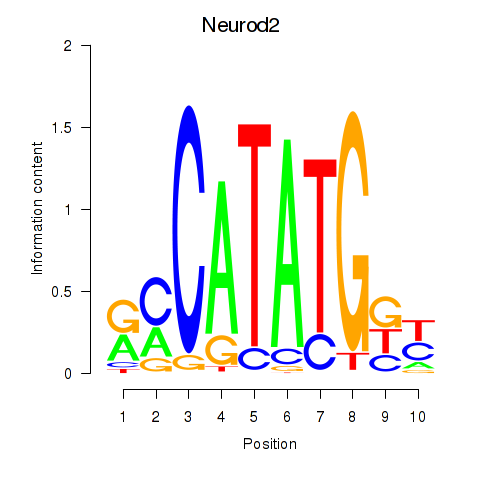
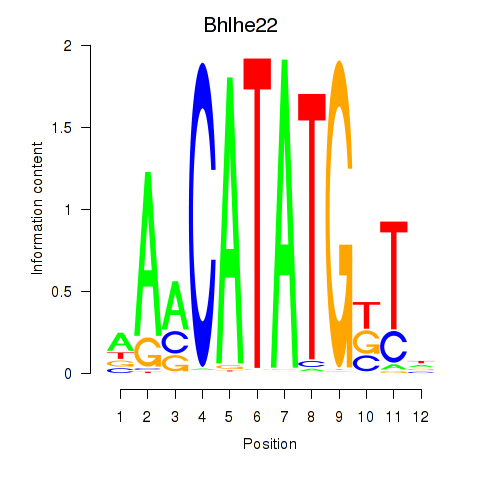
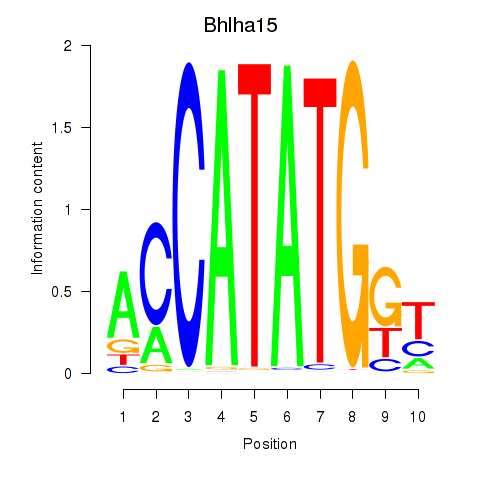
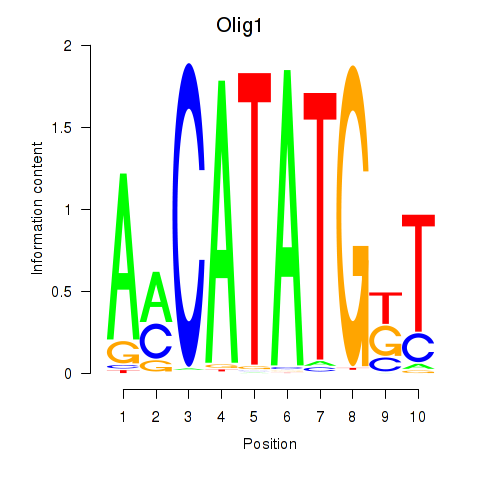
Transcription factors associated with Neurod2_Bhlha15_Bhlhe22_Olig1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bhlha15 | ENSMUSG00000052271.7 | Bhlha15 |
| Bhlhe22 | ENSMUSG00000025128.6 | Bhlhe22 |
| Neurod2 | ENSMUSG00000038255.6 | Neurod2 |
| Olig1 | ENSMUSG00000046160.5 | Olig1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlhe22 | mm10_v2_chr3_+_18054258_18054325 | 0.66 | 2.8e-03 | Click! |
| Neurod2 | mm10_v2_chr11_-_98329641_98329654 | 0.65 | 3.3e-03 | Click! |
| Olig1 | mm10_v2_chr16_+_91269759_91269778 | 0.61 | 6.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.7 | 2.0 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 | 2.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.5 | 2.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.4 | 1.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.3 | 0.9 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 0.6 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 0.2 | 1.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 0.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.2 | 1.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 1.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 1.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.1 | 1.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 3.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 2.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.5 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.3 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:2000097 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) regulation of smooth muscle cell-matrix adhesion(GO:2000097) negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 3.0 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.3 | GO:0061368 | maternal process involved in parturition(GO:0060137) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.8 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0070914 | nucleosome disassembly(GO:0006337) UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 1.1 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.1 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.3 | 2.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.5 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 3.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 4.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 3.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 2.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 2.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.6 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.0 | 1.4 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.0 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME_INTERFERON_GAMMA_SIGNALING | Genes involved in Interferon gamma signaling |