Motif ID: Pbx2
Z-value: 0.777
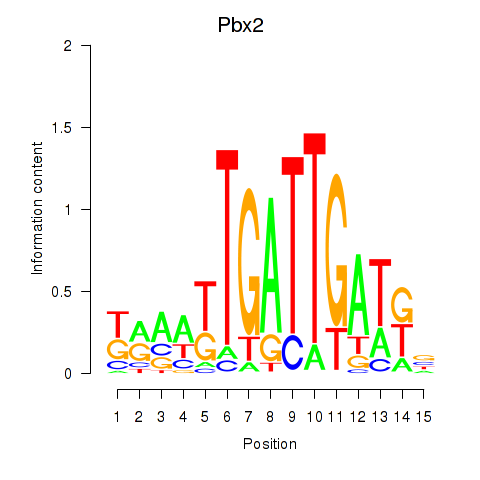
Transcription factors associated with Pbx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pbx2 | ENSMUSG00000034673.8 | Pbx2 |
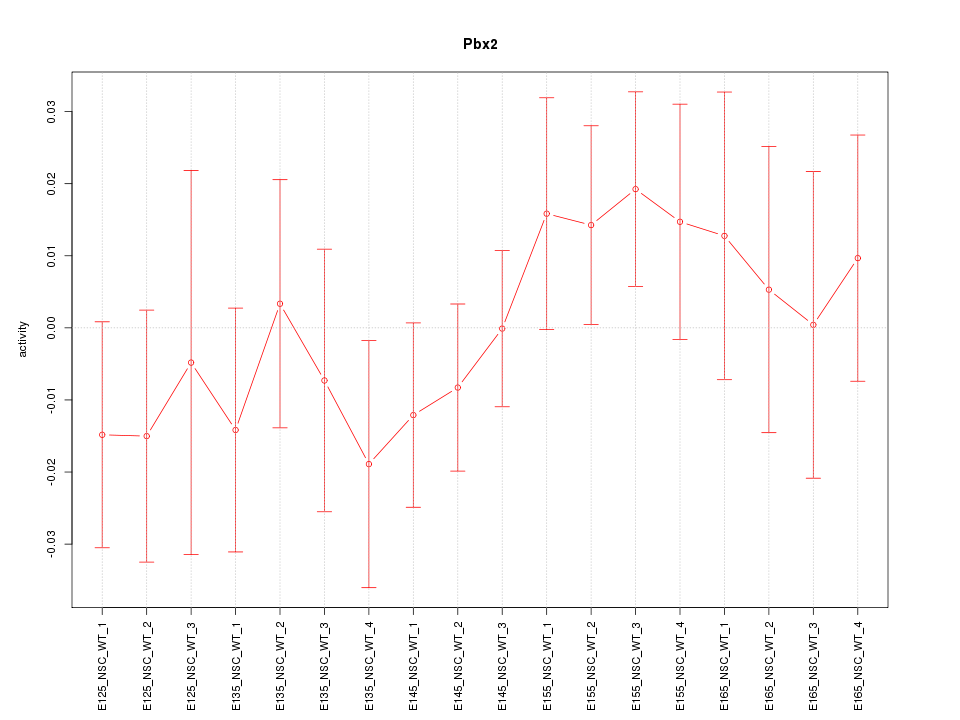
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pbx2 | mm10_v2_chr17_+_34592248_34592329 | -0.79 | 1.1e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.7 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 1.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.3 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 3.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.8 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.0 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 | 0.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.6 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.8 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0051926 | dopamine receptor signaling pathway(GO:0007212) negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 1.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 4.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 1.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 3.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 3.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.1 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.0 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.3 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.9 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.5 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 2.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |