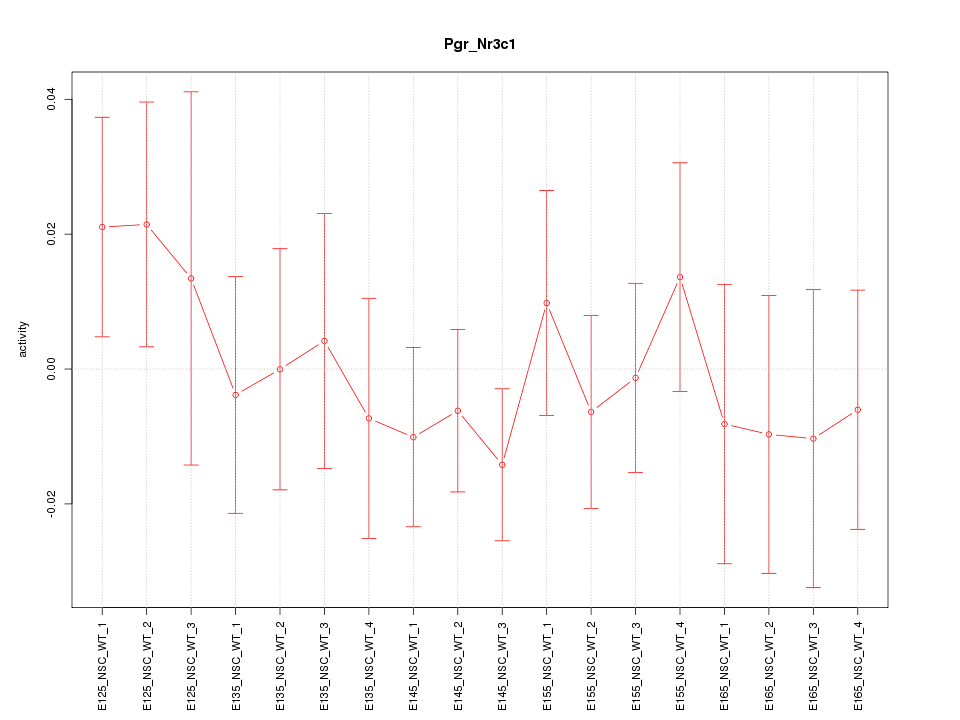
Motif ID: Pgr_Nr3c1
Z-value: 0.663


Transcription factors associated with Pgr_Nr3c1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr3c1 | ENSMUSG00000024431.8 | Nr3c1 |
| Pgr | ENSMUSG00000031870.10 | Pgr |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c1 | mm10_v2_chr18_-_39489776_39489868 | -0.56 | 1.7e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.9 | 3.7 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 1.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.2 | 1.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.2 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 0.5 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.1 | 0.4 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 0.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.5 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.5 | GO:0048386 | positive regulation of keratinocyte differentiation(GO:0045618) positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 | 0.4 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.6 | GO:0021670 | lateral ventricle development(GO:0021670) negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.5 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0046671 | negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.7 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 1.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.7 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.9 | PID_P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 7.3 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.4 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.5 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |