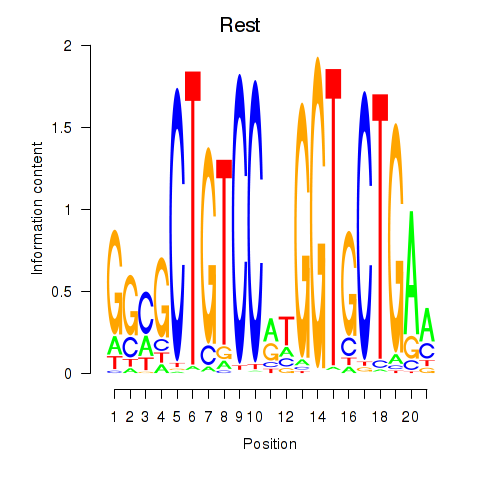
Motif ID: Rest
Z-value: 3.598
Transcription factors associated with Rest:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rest | ENSMUSG00000029249.9 | Rest |
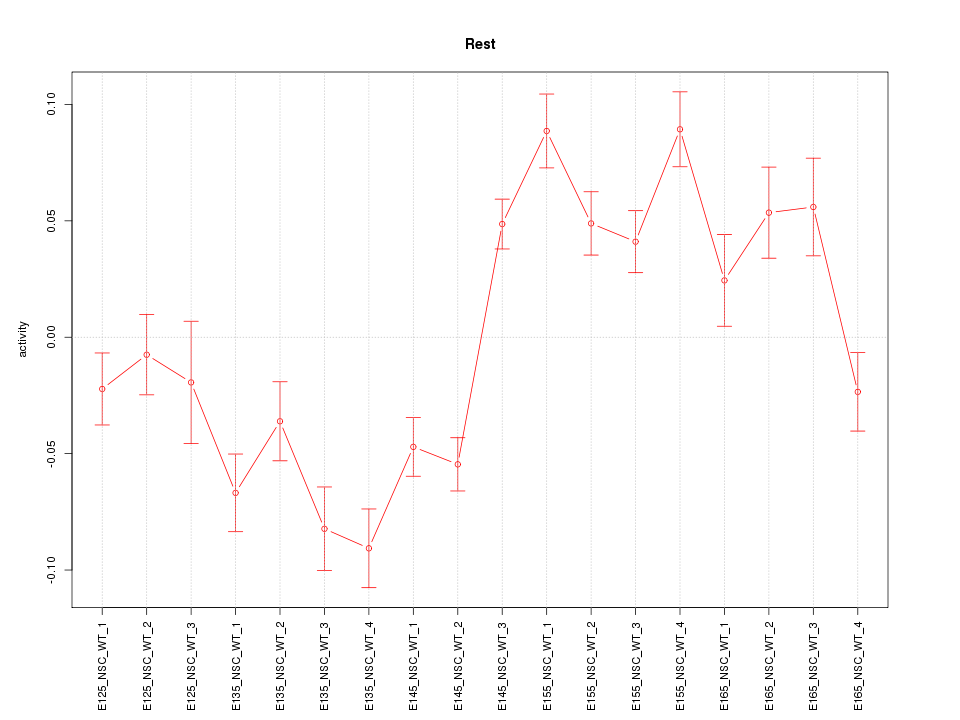
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rest | mm10_v2_chr5_+_77265454_77265549 | -0.70 | 1.2e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.5 | GO:0021586 | pons maturation(GO:0021586) |
| 3.2 | 28.8 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 2.7 | 8.0 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.2 | 10.9 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 2.0 | 18.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.9 | 20.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 1.7 | 6.7 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 1.6 | 9.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.2 | 3.5 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 1.0 | 3.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.0 | 3.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.9 | 9.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.8 | 2.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.8 | 5.0 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.8 | 2.3 | GO:1901079 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) |
| 0.7 | 7.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.7 | 3.9 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.6 | 3.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.6 | 15.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.6 | 6.3 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.5 | 8.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.5 | 13.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 6.0 | GO:0043084 | penile erection(GO:0043084) |
| 0.5 | 10.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.5 | 12.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.5 | 5.9 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.4 | 2.0 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.4 | 5.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.4 | 12.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.4 | 2.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 7.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 2.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.2 | 2.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 3.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 4.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 10.8 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 | 4.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 2.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 3.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.5 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 6.2 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 4.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.4 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 2.5 | 7.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.9 | 28.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.7 | 20.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 1.6 | 9.7 | GO:0008091 | spectrin(GO:0008091) |
| 1.6 | 10.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.8 | 10.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.8 | 10.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.8 | 8.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.7 | 6.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.6 | 2.5 | GO:0002141 | stereocilia ankle link(GO:0002141) |
| 0.6 | 26.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 5.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 18.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 8.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.5 | 2.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 10.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 3.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 16.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 4.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 6.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 8.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 28.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 3.0 | 18.0 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.6 | 12.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.0 | 8.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.6 | 9.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.3 | 12.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 1.0 | 3.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.9 | 2.7 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.8 | 3.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.7 | 20.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.7 | 10.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.6 | 6.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 15.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.5 | 3.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 3.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 13.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 7.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 4.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 9.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 8.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 19.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 5.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 6.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 8.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 2.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 5.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 2.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 4.1 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 0.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.5 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 8.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.8 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.2 | 9.4 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 28.8 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.2 | 7.5 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 6.3 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 47.9 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.1 | 30.8 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 13.9 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.4 | 3.5 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 6.3 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 5.9 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.4 | 7.5 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 5.6 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 0.9 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.6 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 4.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 10.1 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.5 | REACTOME_TRAF6_MEDIATED_INDUCTION_OF_NFKB_AND_MAP_KINASES_UPON_TLR7_8_OR_9_ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.4 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |