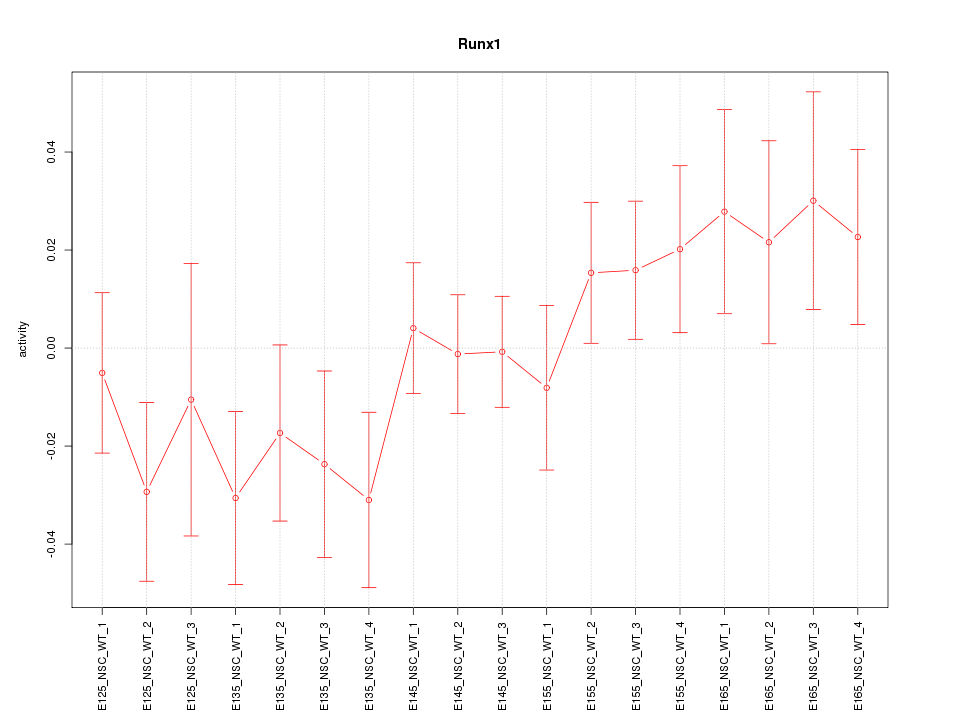
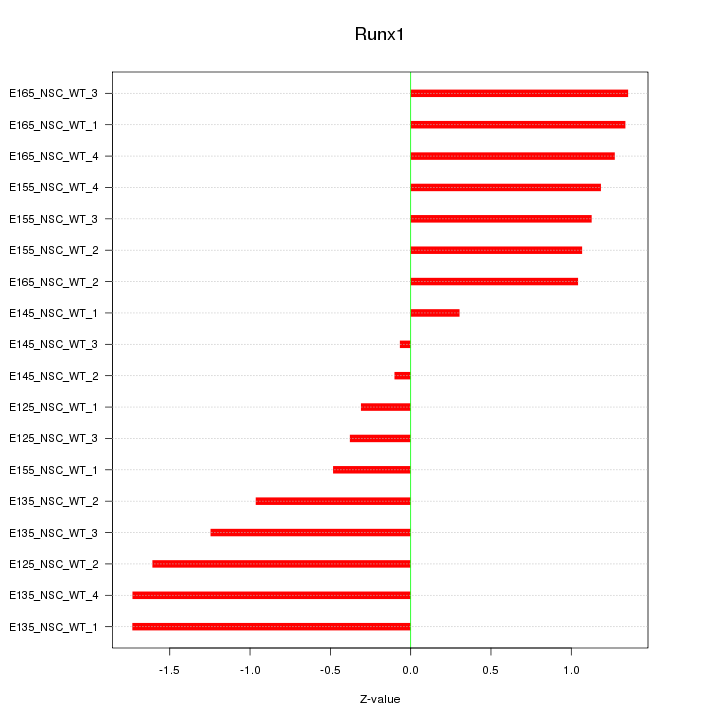
Motif ID: Runx1
Z-value: 1.100
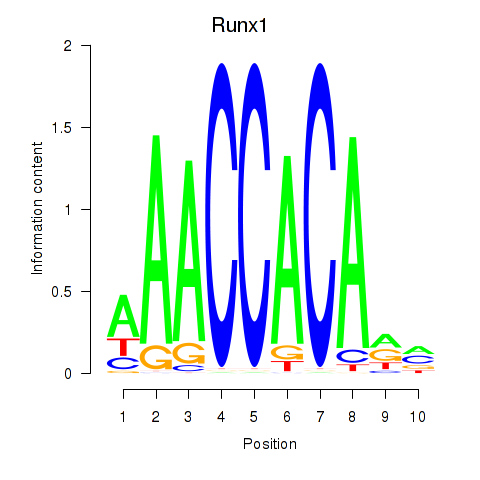
Transcription factors associated with Runx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Runx1 | ENSMUSG00000022952.10 | Runx1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.5 | 2.2 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.4 | 2.2 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.4 | 2.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.4 | 2.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 3.6 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.3 | 2.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 2.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 2.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 1.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.3 | 1.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 1.1 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 | 0.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 2.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.1 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.2 | 1.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 6.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.9 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.1 | 1.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 2.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 8.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.8 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 1.9 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 1.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 1.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.3 | GO:2000318 | positive regulation of interferon-gamma secretion(GO:1902715) positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 3.4 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.1 | 4.1 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.5 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 1.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 1.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.8 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 4.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.7 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 2.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.5 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.5 | GO:0008542 | visual learning(GO:0008542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.5 | 2.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 1.0 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 2.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 2.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 8.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 2.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.3 | 10.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 2.7 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 0.7 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 2.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.5 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 3.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 4.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 4.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 4.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 5.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 2.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 2.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 4.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.6 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.1 | 5.3 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.5 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.5 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.1 | 0.8 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.1 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 8.6 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.9 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.7 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.9 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.0 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 3.6 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 4.7 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.5 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.3 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.2 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.5 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 5.3 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.9 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 2.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.1 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.7 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 2.0 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |