Motif ID: Rxra
Z-value: 0.969
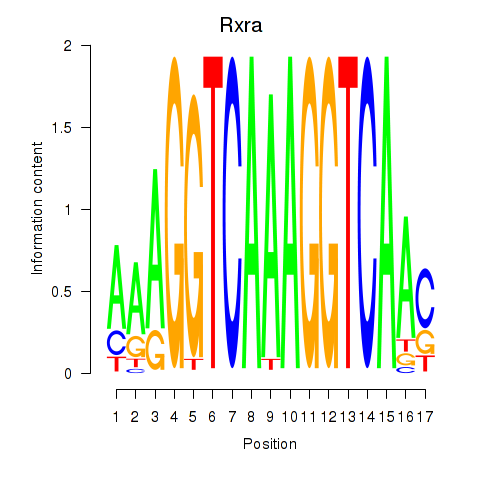
Transcription factors associated with Rxra:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rxra | ENSMUSG00000015846.8 | Rxra |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxra | mm10_v2_chr2_+_27677201_27677229 | -0.66 | 2.8e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.6 | 1.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 3.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.4 | 0.8 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.4 | 1.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 2.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.7 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 0.9 | GO:1904717 | positive regulation of synapse structural plasticity(GO:0051835) regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.2 | 1.5 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 0.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 2.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.8 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.9 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 0.5 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.5 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 2.0 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 1.6 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.6 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.6 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 0.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 1.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.4 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 2.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.0 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.3 | 2.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 1.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 0.8 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.3 | 2.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 1.4 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 0.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.6 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.8 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.3 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) PH domain binding(GO:0042731) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.0 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.4 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.0 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.6 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.6 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.5 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.7 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.2 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |





