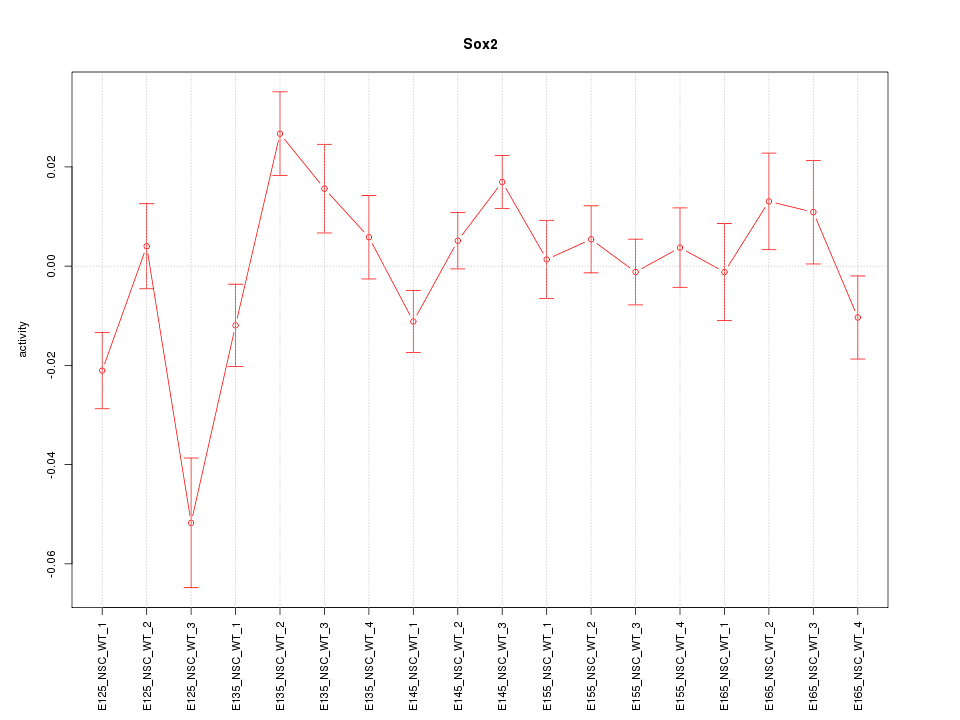
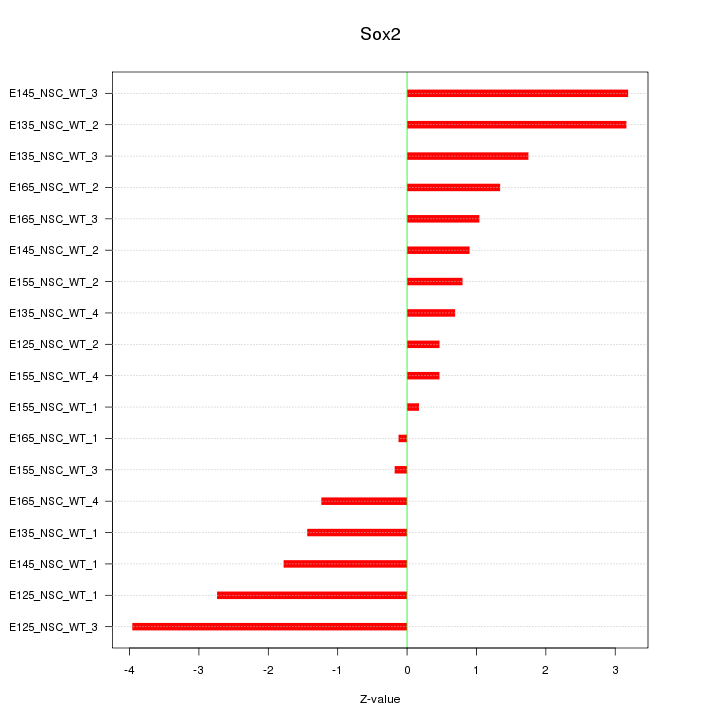
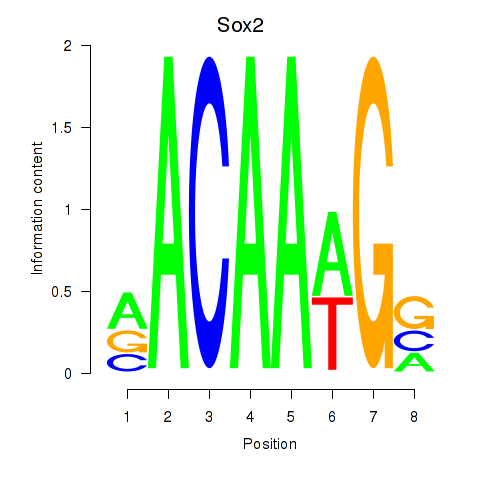
Motif ID: Sox2
Z-value: 1.802
Transcription factors associated with Sox2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox2 | ENSMUSG00000074637.4 | Sox2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox2 | mm10_v2_chr3_+_34649987_34650005 | -0.01 | 9.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.2 | 5.9 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 1.2 | 3.5 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.9 | 3.7 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.9 | 4.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.9 | 2.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.8 | 2.5 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.8 | 3.4 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.8 | 2.4 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.7 | 5.6 | GO:0061368 | maternal process involved in parturition(GO:0060137) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.6 | 2.6 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.6 | 5.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.6 | 1.8 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.6 | 3.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.6 | 2.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.6 | 2.9 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.6 | 2.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.6 | 8.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.6 | 1.7 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.6 | 1.7 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.6 | 2.3 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.6 | 1.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 1.6 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.5 | 1.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.5 | 5.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 1.6 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 0.5 | 2.1 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.5 | 1.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.5 | 1.6 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.5 | 5.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.5 | 2.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 1.5 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.5 | 3.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 | 4.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 | 1.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 1.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.5 | 3.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 4.8 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 1.7 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.4 | 1.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.4 | 3.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 0.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 1.2 | GO:1904207 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.4 | 3.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.4 | 0.8 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.4 | 1.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.4 | 1.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.4 | 3.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.4 | 7.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 2.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.4 | 1.2 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.4 | 0.4 | GO:0061349 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.4 | 1.9 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.4 | 4.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.4 | 2.6 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 2.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 1.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 1.0 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 1.3 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 1.3 | GO:0098904 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) regulation of AV node cell action potential(GO:0098904) |
| 0.3 | 2.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.3 | 1.0 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.3 | 3.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 1.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 0.9 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.3 | 1.2 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 2.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 0.9 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.3 | 1.8 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 0.9 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.3 | 0.9 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.3 | 2.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 0.9 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.3 | 0.6 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.3 | 0.8 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) dorsal root ganglion development(GO:1990791) |
| 0.3 | 1.7 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 | 1.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 0.8 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.3 | 1.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) positive regulation of mesoderm development(GO:2000382) |
| 0.3 | 1.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 0.8 | GO:2000546 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.3 | 1.8 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 0.8 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 1.3 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.2 | 1.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.2 | 0.7 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.2 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 0.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 4.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 1.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 0.7 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 | 0.7 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 0.9 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.2 | 7.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 1.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 0.7 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.2 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.7 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.2 | 0.7 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 0.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.2 | GO:0065001 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) specification of axis polarity(GO:0065001) |
| 0.2 | 0.4 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.2 | 0.4 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.2 | 1.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 2.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.7 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 0.5 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.2 | 0.9 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 1.6 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.5 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.2 | 1.6 | GO:0043615 | astrocyte cell migration(GO:0043615) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 0.5 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 | 0.7 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 0.7 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 0.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.5 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.2 | 0.8 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 0.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.2 | 2.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 1.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 4.7 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 0.9 | GO:1903056 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 0.8 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.2 | 2.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.2 | 0.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 1.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.5 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
| 0.1 | 0.4 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.1 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.6 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.9 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 6.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.7 | GO:0018377 | N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 | 0.7 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.4 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.6 | GO:1904528 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 0.5 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.7 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 1.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 3.4 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 1.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 2.1 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 0.7 | GO:0036438 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.5 | GO:1900164 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.1 | 1.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 3.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 2.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 2.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.2 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.1 | 0.5 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.8 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.1 | 1.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.9 | GO:0021889 | olfactory bulb interneuron differentiation(GO:0021889) |
| 0.1 | 1.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 1.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.5 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.4 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.5 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.1 | 0.8 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.9 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 1.2 | GO:1903170 | negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.1 | 1.0 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.9 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.3 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 3.1 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.5 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.7 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.4 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.5 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.0 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 1.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 2.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.8 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.2 | GO:0090646 | mitochondrial RNA 3'-end processing(GO:0000965) mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 2.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.5 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.5 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0051133 | regulation of NK T cell activation(GO:0051133) positive regulation of NK T cell activation(GO:0051135) |
| 0.1 | 0.5 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.1 | 0.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 4.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 1.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 0.5 | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation(GO:0010998) regulation of translational initiation in response to stress(GO:0043558) |
| 0.1 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.8 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 0.8 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 3.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 2.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.0 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 1.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.3 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.0 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 1.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.9 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.1 | 0.7 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 1.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 2.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 1.3 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.7 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.3 | GO:0046364 | hexose biosynthetic process(GO:0019319) monosaccharide biosynthetic process(GO:0046364) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.5 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 0.3 | GO:0007440 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 1.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0060353 | cell adhesion molecule production(GO:0060352) regulation of cell adhesion molecule production(GO:0060353) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 2.4 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 1.4 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) cellular response to endothelin(GO:1990859) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.9 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.4 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.0 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 1.4 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0072176 | nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.9 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.3 | GO:0015844 | monoamine transport(GO:0015844) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0071442 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:2000483 | detection of bacterium(GO:0016045) detection of molecule of bacterial origin(GO:0032490) response to bacterial lipoprotein(GO:0032493) detection of other organism(GO:0098543) negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.6 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:1904008 | cellular response to salt(GO:1902075) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.1 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.3 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 1.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.0 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 | 0.3 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0061097 | regulation of protein tyrosine kinase activity(GO:0061097) |
| 0.0 | 0.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.5 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 1.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:1903901 | negative regulation of viral process(GO:0048525) negative regulation of viral life cycle(GO:1903901) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.0 | GO:2000744 | midbrain-hindbrain boundary development(GO:0030917) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.4 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.9 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0001947 | heart looping(GO:0001947) determination of heart left/right asymmetry(GO:0061371) |
| 0.0 | 0.2 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.7 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.0 | 0.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.3 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 1.0 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 3.3 | GO:0006820 | anion transport(GO:0006820) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.9 | 12.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.8 | 3.4 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.8 | 7.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 2.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.6 | 7.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.5 | 2.1 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.5 | 2.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 1.4 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.5 | 1.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 1.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.4 | 1.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.3 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 1.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 1.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 1.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.3 | 1.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 4.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 5.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 3.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 2.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 1.8 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.2 | 1.8 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 0.6 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.2 | 1.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 4.2 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 1.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.7 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.4 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 8.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 4.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 2.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 2.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 2.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.3 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 1.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 3.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 3.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 1.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.0 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 3.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 2.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 12.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0043256 | laminin-1 complex(GO:0005606) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 2.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 3.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 1.0 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 2.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 3.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 10.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.9 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 1.0 | 8.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.8 | 5.7 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.7 | 3.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.7 | 2.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.6 | 4.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.6 | 3.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.6 | 2.4 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.5 | 3.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 3.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 2.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 3.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 3.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.4 | 3.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 3.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.4 | 1.2 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.4 | 5.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 2.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 3.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 1.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.3 | 1.0 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.3 | 1.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.3 | 1.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 0.9 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 7.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 0.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 1.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 0.8 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.3 | 15.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 1.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 0.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.3 | 0.8 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.3 | 0.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 2.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 0.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 1.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 2.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 6.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 3.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 9.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 2.5 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.2 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 3.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.5 | GO:0005168 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 1.0 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.6 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.2 | 2.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 4.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 0.5 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.1 | 2.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.2 | GO:0070325 | lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.7 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 2.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.7 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.4 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 1.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 2.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.4 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 7.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 7.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 0.3 | GO:0004534 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.3 | GO:0070699 | activin receptor binding(GO:0070697) type II activin receptor binding(GO:0070699) |
| 0.1 | 18.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.4 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 0.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 3.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 6.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 3.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.7 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 1.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 3.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 11.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 3.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 3.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0005416 | sodium:amino acid symporter activity(GO:0005283) cation:amino acid symporter activity(GO:0005416) |
| 0.0 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 7.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 3.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 2.0 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 7.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 4.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 14.2 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 5.4 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.1 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.2 | 12.8 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.0 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.8 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 6.7 | PID_ERBB1_RECEPTOR_PROXIMAL_PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 1.9 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.3 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.5 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 0.3 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 2.7 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 2.5 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 5.2 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 0.8 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.6 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.8 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.6 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.2 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 1.1 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.7 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 3.7 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.4 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.1 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.8 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.1 | 3.8 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.2 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.5 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.8 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.6 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.3 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 0.9 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 3.3 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.8 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 2.0 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.2 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 1.0 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 2.9 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.1 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.3 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.4 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 1.8 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 5.4 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 1.8 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 5.6 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 1.6 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 4.4 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 13.6 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.2 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 2.2 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.3 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.3 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.5 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 4.8 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.7 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.1 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.1 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.5 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 3.9 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.7 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 10.4 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 3.0 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.9 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.1 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.1 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.5 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 5.0 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.5 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.9 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.7 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.2 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.0 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.5 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.3 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.7 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.8 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 5.2 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.3 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.8 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.4 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.6 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.7 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.8 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.5 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.1 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.3 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.0 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.4 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.7 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.8 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME_CHONDROITIN_SULFATE_DERMATAN_SULFATE_METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.8 | REACTOME_PI_METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.5 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.1 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME_MRNA_CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.4 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.8 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.0 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |