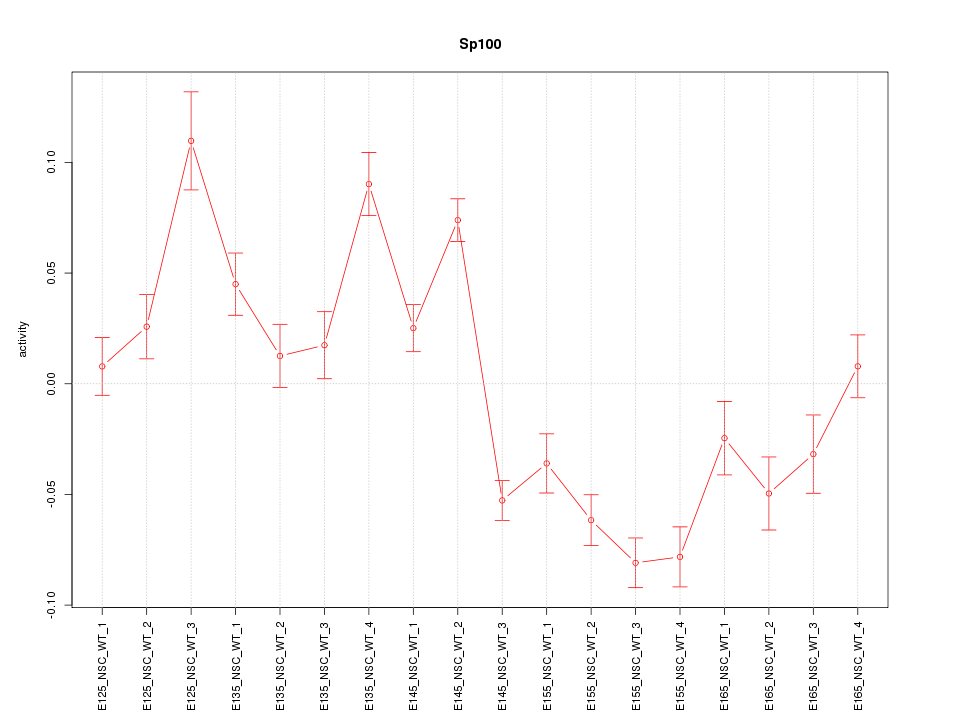
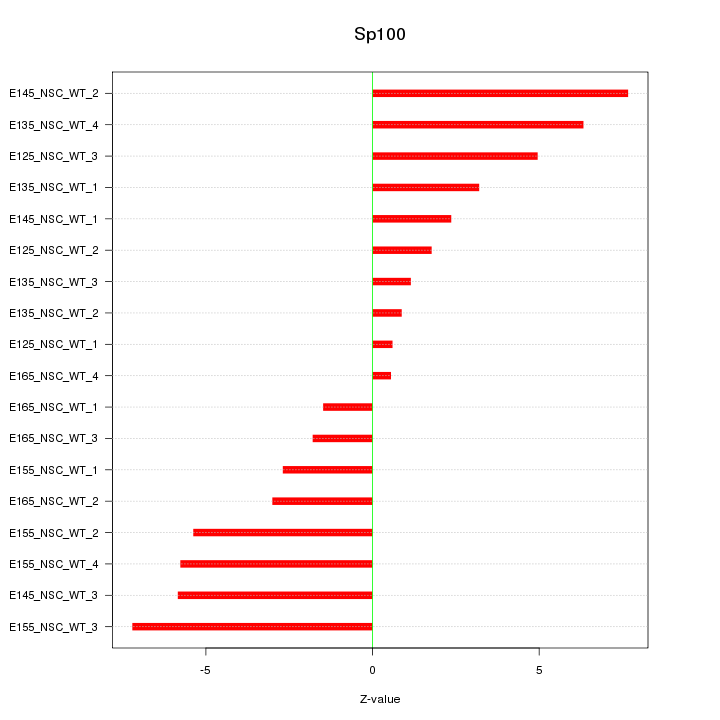
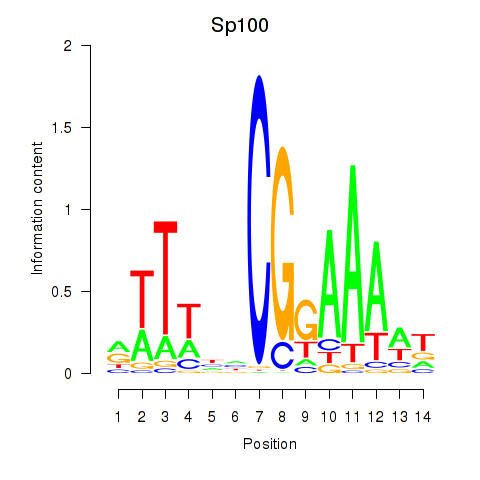
Motif ID: Sp100
Z-value: 4.184
Transcription factors associated with Sp100:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sp100 | ENSMUSG00000026222.10 | Sp100 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm10_v2_chr1_+_85650008_85650020 | -0.21 | 4.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.1 | 5.5 | GO:0021764 | amygdala development(GO:0021764) |
| 1.0 | 3.1 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.9 | 5.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.8 | 5.8 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 0.7 | 5.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.7 | 2.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.7 | 4.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.7 | 7.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.7 | 12.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 1.9 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.6 | 18.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 4.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.6 | 1.7 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.6 | 2.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.4 | 2.7 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.4 | 1.6 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.4 | 1.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.4 | 1.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.4 | 2.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.4 | 4.0 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.3 | 7.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.3 | 2.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 3.9 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 2.9 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 0.9 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.3 | 1.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.3 | 2.5 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 1.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 0.8 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.2 | 4.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.2 | 2.5 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 | 1.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 1.2 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.2 | 1.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 7.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 0.5 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 6.9 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 1.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 2.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.2 | 0.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 3.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 2.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 1.8 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 1.4 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.2 | 0.6 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 1.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 1.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 4.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.7 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 3.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 2.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.7 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 2.9 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 1.9 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.1 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.1 | 1.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.3 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 1.4 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 2.0 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.8 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 2.6 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 6.3 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 1.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.0 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.1 | 2.3 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 4.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 8.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 3.6 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.1 | 0.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 3.0 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 2.0 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 1.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.4 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 1.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.5 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 5.2 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.9 | 5.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.8 | 2.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.7 | 2.8 | GO:0043293 | apoptosome(GO:0043293) |
| 0.6 | 18.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.5 | 4.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.4 | 5.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 2.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.4 | 3.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.4 | 3.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 3.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.4 | 3.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 2.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 1.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 1.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 1.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 4.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 5.7 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 3.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 5.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 2.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 3.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 2.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 5.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.6 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 6.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 1.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 2.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 5.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 4.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) replication fork(GO:0005657) |
| 0.1 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.2 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 3.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.0 | 3.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 1.0 | 7.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.9 | 4.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.8 | 3.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.7 | 3.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.6 | 4.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.6 | 3.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 4.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 7.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.4 | 2.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.4 | 1.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.3 | 3.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 2.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 1.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.3 | 2.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 2.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 2.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 0.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 12.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 10.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 0.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.7 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.2 | 1.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 1.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 0.5 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 4.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 3.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 1.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 5.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 2.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 5.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 4.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 5.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 9.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.7 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 5.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.5 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 8.0 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 3.8 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 5.5 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.3 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.4 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.7 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.1 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.0 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 1.1 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.1 | 0.7 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.8 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.2 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.2 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.9 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.6 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.2 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 4.5 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 20.3 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 2.9 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 3.1 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.6 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 1.8 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.5 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.3 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.0 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.5 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.9 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.9 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.7 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 6.1 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 2.6 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 5.3 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.6 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 5.9 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.5 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.4 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 5.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.5 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.3 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 3.4 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 5.2 | REACTOME_DIABETES_PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 2.3 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.2 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 4.2 | REACTOME_3_UTR_MEDIATED_TRANSLATIONAL_REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 1.2 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.4 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |