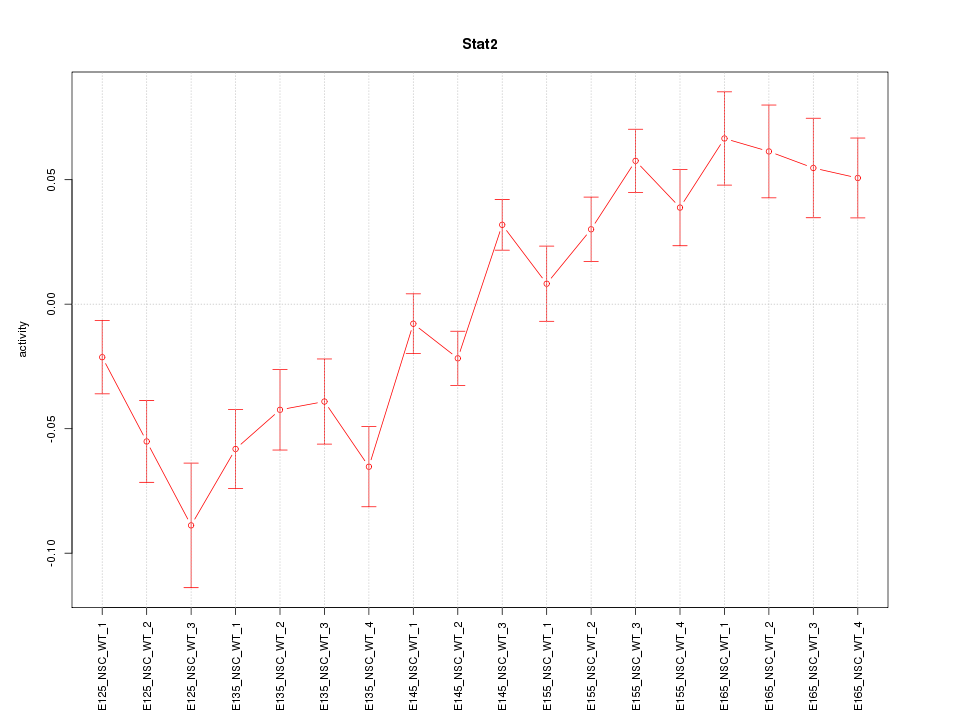
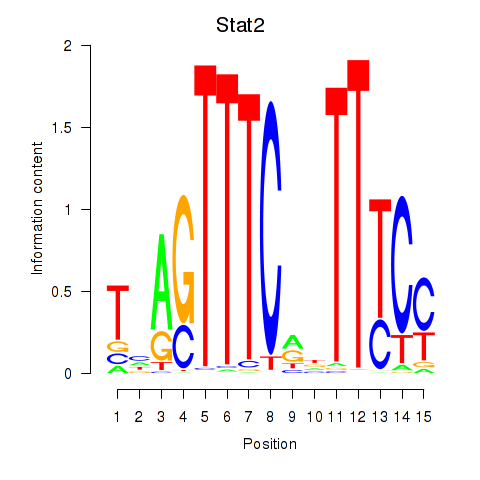
Motif ID: Stat2
Z-value: 2.943
Transcription factors associated with Stat2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Stat2 | ENSMUSG00000040033.9 | Stat2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat2 | mm10_v2_chr10_+_128270546_128270577 | 0.86 | 5.8e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 4.3 | 13.0 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 2.0 | 5.9 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 1.9 | 5.7 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.8 | 5.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 1.7 | 6.7 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.6 | 28.0 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 1.5 | 6.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 1.5 | 4.5 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 1.4 | 4.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 1.4 | 4.3 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 1.4 | 4.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 1.2 | 5.9 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.1 | 6.8 | GO:0032439 | endosome localization(GO:0032439) |
| 1.0 | 1.0 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) regulation of antigen processing and presentation of peptide antigen(GO:0002583) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.0 | 3.0 | GO:0002925 | regulation of dendritic cell cytokine production(GO:0002730) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) detection of peptidoglycan(GO:0032499) negative regulation of interleukin-18 production(GO:0032701) |
| 1.0 | 4.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.9 | 3.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.9 | 2.7 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.9 | 2.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.8 | 5.1 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 0.8 | 6.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.7 | 12.4 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.7 | 4.3 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.7 | 4.3 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.7 | 1.4 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.7 | 2.8 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.7 | 7.0 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.7 | 5.4 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.7 | 3.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.7 | 24.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.7 | 2.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.6 | 16.4 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.6 | 1.9 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid metabolic process(GO:0046416) |
| 0.6 | 3.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.6 | 3.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.5 | 1.6 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.5 | 2.7 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.5 | 1.6 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.5 | 1.5 | GO:1902915 | negative regulation of protein import into nucleus, translocation(GO:0033159) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 2.9 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.5 | 2.9 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.5 | 4.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 1.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.4 | 1.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 6.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.4 | 4.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 2.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 1.2 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) quinolinate biosynthetic process(GO:0019805) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.4 | 10.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.3 | 3.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 2.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 4.2 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 0.9 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 2.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 2.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 3.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 18.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.3 | 1.9 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 1.8 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 4.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 11.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 1.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 3.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.2 | 1.5 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.2 | 0.8 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.2 | 3.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.2 | 0.4 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.2 | 2.6 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.2 | 1.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 1.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 2.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 1.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 3.8 | GO:0043473 | pigmentation(GO:0043473) |
| 0.1 | 2.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 4.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 1.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 1.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.6 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.7 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 4.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 7.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.5 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 3.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 2.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 2.4 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 1.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.3 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 3.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) regulation of somitogenesis(GO:0014807) |
| 0.0 | 2.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 3.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.7 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 1.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 2.1 | GO:0003015 | heart process(GO:0003015) |
| 0.0 | 0.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 7.2 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 1.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.0 | 3.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.9 | 5.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.9 | 4.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 11.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 6.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.7 | 2.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.6 | 3.9 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.5 | 2.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 2.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 4.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 3.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.5 | 2.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 2.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 1.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 2.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 28.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 4.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 2.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 7.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 1.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 22.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 3.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 2.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 4.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 12.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 10.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 20.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 8.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0042175 | endoplasmic reticulum membrane(GO:0005789) nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 24.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 8.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 9.7 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 2.0 | 5.9 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) |
| 1.7 | 6.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.6 | 4.9 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 1.6 | 11.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.3 | 5.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.1 | 4.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 1.1 | 4.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.1 | 4.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.9 | 4.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.9 | 2.7 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.9 | 2.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.9 | 2.6 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.8 | 4.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.8 | 3.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.7 | 3.0 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.7 | 6.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 6.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.6 | 4.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.5 | 18.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.5 | 2.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 5.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.4 | 2.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 2.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 1.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.4 | 1.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 4.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.4 | 28.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.3 | 1.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 7.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 3.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 2.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 2.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 4.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 6.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 5.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 4.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 4.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 0.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 7.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 2.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 33.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 1.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 2.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 4.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 8.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 4.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 6.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 8.4 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 1.4 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 12.0 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.1 | 5.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 2.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 8.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 4.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 23.0 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.6 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.3 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 2.7 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.7 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 6.7 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.3 | 2.6 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 4.1 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.2 | 3.0 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.4 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.1 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.3 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 19.4 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 5.4 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.5 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.1 | 7.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 2.1 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 3.4 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.5 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 1.6 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.1 | 4.3 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.7 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.3 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 49.9 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.6 | 4.7 | REACTOME_PD1_SIGNALING | Genes involved in PD-1 signaling |
| 0.5 | 11.2 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 4.5 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 7.8 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 6.4 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 8.4 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 6.7 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.2 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.4 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.3 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.7 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 4.3 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.0 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 2.3 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.6 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.6 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 4.2 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 2.7 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.1 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.9 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.3 | REACTOME_TOLL_RECEPTOR_CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 2.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |