Motif ID: Tbx15
Z-value: 1.039
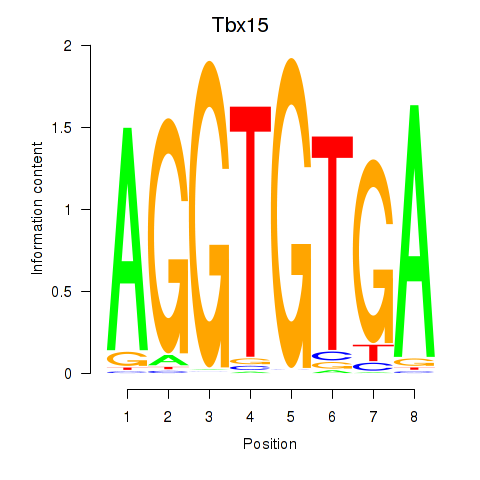
Transcription factors associated with Tbx15:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbx15 | ENSMUSG00000027868.5 | Tbx15 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.9 | 1.9 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.3 | 4.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 1.2 | 2.3 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 1.1 | 3.3 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 1.1 | 3.3 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.0 | 3.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.8 | 2.3 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.7 | 3.6 | GO:0098735 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.7 | 2.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.7 | 2.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 | 3.7 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.6 | 4.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.5 | 4.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 2.2 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.5 | 2.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.5 | 1.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.4 | 1.3 | GO:1904580 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) regulation of intracellular mRNA localization(GO:1904580) |
| 0.4 | 1.3 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.3 | 1.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.3 | 1.7 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 1.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 1.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 | 0.9 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 | 0.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 | 1.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.3 | 1.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 1.4 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.2 | 0.9 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.2 | 0.7 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 1.6 | GO:0098909 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.2 | 1.7 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 2.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.2 | 1.9 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 1.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 1.6 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 0.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.2 | 0.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 3.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 0.7 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.2 | 0.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 1.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 0.5 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.2 | 0.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 3.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.6 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.6 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 1.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.7 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 2.0 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.7 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 1.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 1.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.2 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 2.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.4 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.1 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.3 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.4 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.3 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 1.1 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 2.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.8 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 2.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 0.9 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.8 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.5 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.4 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.4 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 0.4 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 2.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.6 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 2.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.3 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 1.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.4 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.4 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 1.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.0 | 0.6 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 2.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.3 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 1.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.3 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 1.3 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.0 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.4 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.4 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.0 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.8 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.5 | 0.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.5 | 3.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 1.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 3.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 1.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 1.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 2.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 3.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 2.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 2.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 5.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 5.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 3.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.4 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 1.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 3.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 6.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 3.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.4 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.9 | 2.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.9 | 3.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.7 | 2.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.7 | 3.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.6 | 2.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.6 | 4.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.6 | 1.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 2.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.5 | 1.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 1.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 1.0 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 1.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 1.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.3 | 5.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.3 | 0.9 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 2.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 2.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.3 | 3.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 2.0 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.3 | 1.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.9 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 0.9 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 1.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 0.5 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 5.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.9 | GO:0044213 | intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 1.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.3 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.9 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.7 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 1.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 3.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.0 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 0.7 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.4 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 3.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.2 | 3.7 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.2 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.1 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.9 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.3 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 1.3 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.3 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.1 | 0.8 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.2 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.0 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.2 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.3 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.7 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 3.4 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID_ATR_PATHWAY | ATR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.2 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 3.6 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 5.0 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.0 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 3.1 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.1 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.9 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.3 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.6 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.4 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.2 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.3 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.2 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 0.9 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.2 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.6 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.5 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.7 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.1 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 3.1 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME_CTLA4_INHIBITORY_SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME_TRIGLYCERIDE_BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |





