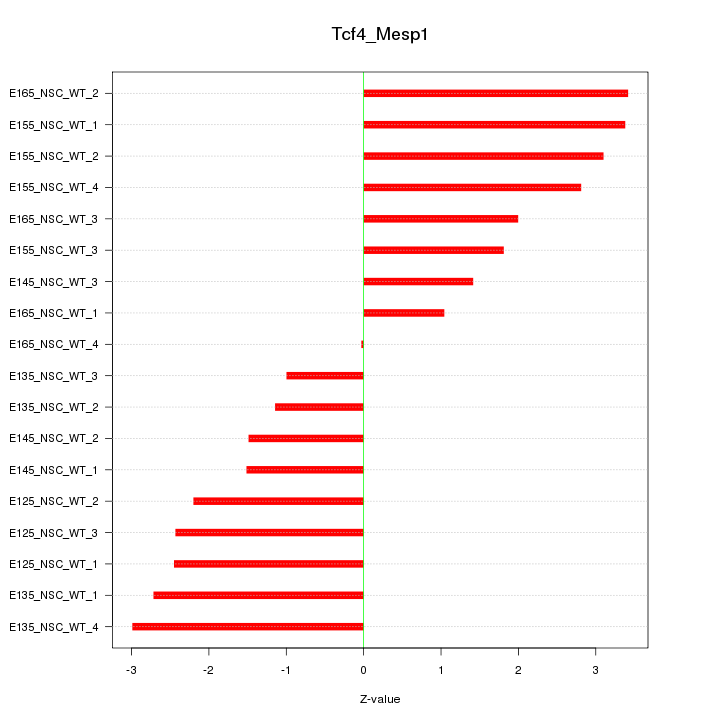
Motif ID: Tcf4_Mesp1
Z-value: 2.247

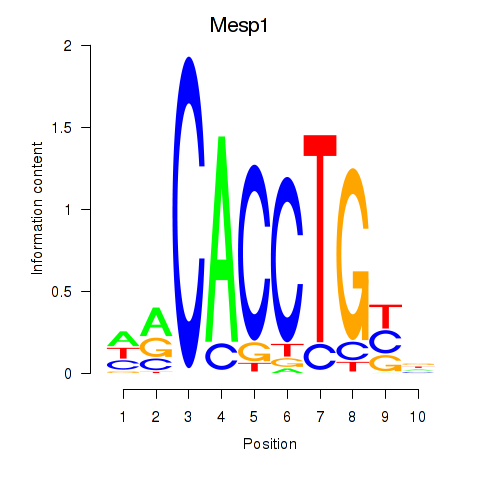
Transcription factors associated with Tcf4_Mesp1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mesp1 | ENSMUSG00000030544.5 | Mesp1 |
| Tcf4 | ENSMUSG00000053477.9 | Tcf4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf4 | mm10_v2_chr18_+_69593361_69593450 | 0.74 | 4.4e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 2.2 | 6.7 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.9 | 5.8 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.7 | 5.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.7 | 6.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.5 | 4.4 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.3 | 2.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 1.3 | 12.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 1.2 | 5.0 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 1.2 | 3.6 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.1 | 3.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.1 | 3.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.0 | 3.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 1.0 | 4.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.0 | 4.1 | GO:2000832 | androgen secretion(GO:0035935) testosterone secretion(GO:0035936) negative regulation of steroid hormone secretion(GO:2000832) regulation of androgen secretion(GO:2000834) regulation of testosterone secretion(GO:2000843) |
| 1.0 | 3.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 1.0 | 2.9 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.0 | 5.9 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.9 | 1.8 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.9 | 2.7 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.9 | 2.7 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.9 | 7.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.9 | 3.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.9 | 4.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.9 | 1.8 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.9 | 4.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.9 | 7.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.9 | 4.3 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.8 | 2.5 | GO:0032430 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.8 | 1.6 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.7 | 3.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.7 | 8.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.7 | 7.0 | GO:0046959 | habituation(GO:0046959) |
| 0.7 | 12.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.7 | 2.7 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.7 | 2.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.7 | 2.0 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.7 | 4.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 2.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.7 | 3.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.7 | 2.0 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.6 | 3.8 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.6 | 3.1 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.6 | 4.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.6 | 6.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.6 | 1.8 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.6 | 4.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.6 | 11.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.6 | 4.1 | GO:2000662 | interleukin-5 secretion(GO:0072603) interleukin-13 secretion(GO:0072611) regulation of interleukin-5 secretion(GO:2000662) regulation of interleukin-13 secretion(GO:2000665) |
| 0.6 | 1.8 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.6 | 1.7 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.6 | 2.3 | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.6 | 1.7 | GO:0015866 | ADP transport(GO:0015866) |
| 0.6 | 1.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.6 | 2.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 6.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.5 | 5.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.5 | 1.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.5 | 0.5 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.5 | 2.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 3.6 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.5 | 1.5 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.5 | 6.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.5 | 1.5 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.5 | 1.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.5 | 6.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.5 | 2.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.5 | 2.9 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.5 | 1.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.5 | 4.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.5 | 2.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.5 | 0.9 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.5 | 1.9 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.5 | 1.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.5 | 6.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 0.9 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.4 | 3.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.4 | 5.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 0.4 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) |
| 0.4 | 1.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.4 | 1.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 9.6 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.4 | 1.2 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 1.6 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.4 | 3.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.4 | 0.8 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.4 | 0.4 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.4 | 0.8 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.4 | 1.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.4 | 0.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.4 | 1.1 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) regulation of bleb assembly(GO:1904170) |
| 0.4 | 1.1 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.4 | 1.1 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.4 | 1.1 | GO:0043379 | rRNA export from nucleus(GO:0006407) memory T cell differentiation(GO:0043379) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.4 | 0.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.4 | 1.9 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.4 | 1.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.4 | 1.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.4 | 1.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.4 | 3.3 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.4 | 1.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.4 | 0.7 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.4 | 1.8 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.4 | 0.4 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.4 | 1.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.4 | 9.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 1.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 | 0.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.3 | 2.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.0 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.3 | 7.0 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.3 | 0.7 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.3 | 1.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.3 | 0.7 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.3 | 1.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 3.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.3 | 1.0 | GO:0072720 | cellular response to UV-A(GO:0071492) response to dithiothreitol(GO:0072720) |
| 0.3 | 2.8 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.3 | 1.2 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 | 1.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 0.9 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.3 | 4.5 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 1.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 1.5 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.3 | 1.2 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 | 1.5 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.3 | 1.8 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.3 | 1.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 3.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.3 | 2.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.3 | 0.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 0.9 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.3 | 0.8 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.3 | 3.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 | 1.1 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.3 | 0.8 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 1.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 0.8 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.3 | 1.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.3 | 0.8 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.3 | 7.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.6 | GO:1902805 | positive regulation of synaptic vesicle transport(GO:1902805) |
| 0.3 | 5.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 1.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.3 | 0.5 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of cortisol secretion(GO:0051464) |
| 0.3 | 1.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.3 | 1.3 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.3 | 1.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.3 | 0.5 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.2 | 1.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.5 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 2.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 1.4 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.2 | 5.5 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 0.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 1.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 0.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 1.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.7 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.2 | 0.7 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.2 | 0.5 | GO:0031622 | positive regulation of fever generation(GO:0031622) |
| 0.2 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.7 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 0.2 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.2 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.2 | 0.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 0.7 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.2 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.9 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 2.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.2 | 13.4 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.2 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.9 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.2 | 3.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 0.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 0.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 1.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 1.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 1.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 1.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.2 | 0.4 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.2 | 1.4 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 0.8 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 0.8 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 0.6 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of vascular wound healing(GO:0061043) |
| 0.2 | 0.6 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 0.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 0.6 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.2 | 0.4 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.2 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 0.6 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.2 | 0.8 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 1.4 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.2 | 1.4 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.2 | 3.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 3.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.4 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.2 | 2.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 1.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 1.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 1.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 3.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 0.6 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.2 | 1.3 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.2 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 3.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.7 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 | 7.7 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.2 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 1.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.5 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.2 | 1.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 6.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 0.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.8 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.2 | 0.5 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 0.9 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 0.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 2.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 2.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 1.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 2.2 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 | 2.5 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.2 | 8.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 0.8 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.2 | 0.5 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.2 | 0.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 0.7 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 1.0 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.2 | 3.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 2.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 0.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 2.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.2 | 0.8 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 0.6 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.2 | 2.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 0.6 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.2 | 6.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 5.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 0.3 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.2 | 0.5 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.2 | 1.6 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.2 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 3.3 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.2 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.8 | GO:0032796 | uropod organization(GO:0032796) |
| 0.2 | 1.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 | 0.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 0.2 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 8.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 1.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.6 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 1.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.6 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.9 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.1 | 2.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.3 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.0 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 0.3 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 1.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.4 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.1 | 1.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.6 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 3.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.6 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 1.7 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 1.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.6 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 0.5 | GO:0090335 | regulation of brown fat cell differentiation(GO:0090335) positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.3 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.5 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.7 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.7 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.1 | 1.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.3 | GO:1903320 | regulation of protein modification by small protein conjugation or removal(GO:1903320) |
| 0.1 | 1.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 2.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 3.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.7 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 3.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.0 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.7 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 1.6 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.4 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.1 | 0.3 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 | 8.7 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 1.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.7 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.5 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.1 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 1.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.5 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.5 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) cellular response to cold(GO:0070417) |
| 0.1 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.8 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 2.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.5 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 1.5 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 1.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 2.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.6 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.8 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 6.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 4.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.5 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 1.8 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.6 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.7 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.3 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.1 | 1.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 9.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.3 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 7.8 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.9 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.2 | GO:0031620 | fever generation(GO:0001660) regulation of fever generation(GO:0031620) |
| 0.1 | 0.9 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.1 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.4 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 1.0 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.0 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 4.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.3 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.1 | 0.3 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 1.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 0.6 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.2 | GO:0019732 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) regulation of defense response to fungus(GO:1900150) regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.1 | 0.7 | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901028) |
| 0.1 | 0.3 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.1 | 0.1 | GO:0060126 | hypophysis morphogenesis(GO:0048850) somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 | 0.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 0.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.6 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.1 | 0.5 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.9 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 1.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.3 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 7.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 1.6 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.1 | 0.2 | GO:0051956 | negative regulation of amino acid transport(GO:0051956) |
| 0.1 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.6 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 2.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 2.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.0 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.1 | 0.7 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.1 | GO:2000561 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) negative regulation of alpha-beta T cell proliferation(GO:0046642) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 0.2 | GO:0048557 | embryonic digestive tract morphogenesis(GO:0048557) |
| 0.1 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 0.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 1.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 3.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 9.0 | GO:0050890 | cognition(GO:0050890) |
| 0.1 | 0.7 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.5 | GO:0046545 | development of primary female sexual characteristics(GO:0046545) |
| 0.1 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.9 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.9 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.3 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) progesterone metabolic process(GO:0042448) |
| 0.0 | 1.0 | GO:1901216 | positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 2.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.3 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.1 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 2.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 1.1 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.8 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 3.6 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 2.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.9 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.5 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.2 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:1901727 | regulation of histone deacetylase activity(GO:1901725) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.6 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 0.2 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 1.7 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.2 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.2 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.0 | GO:0060768 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.6 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.0 | 0.2 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0043512 | inhibin A complex(GO:0043512) |
| 1.3 | 3.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.1 | 4.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.0 | 4.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.0 | 3.0 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.9 | 5.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.9 | 1.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.9 | 3.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.7 | 9.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.7 | 2.2 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.7 | 7.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.7 | 2.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.7 | 10.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.7 | 3.4 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.6 | 17.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.6 | 1.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.6 | 4.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 5.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 2.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 2.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.5 | 1.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.5 | 4.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 7.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 4.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 5.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 1.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.4 | 1.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.4 | 3.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 1.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.4 | 1.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 2.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.3 | 3.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 8.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 4.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 2.8 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 2.5 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 1.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 42.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.3 | 11.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 1.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 4.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 6.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 1.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 1.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 11.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 1.5 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.3 | 3.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 5.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 0.7 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 2.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 14.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 2.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 3.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 2.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 2.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 3.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 1.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 8.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 1.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 1.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 0.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.2 | 0.8 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 0.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 2.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 42.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 14.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 17.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 3.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 7.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.3 | GO:0044754 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.1 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 18.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 3.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 2.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 1.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 2.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.3 | GO:1902636 | kinocilium(GO:0060091) kinociliary basal body(GO:1902636) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 7.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 2.0 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 4.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 1.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 7.5 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 3.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.7 | GO:0071004 | U1 snRNP(GO:0005685) U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 4.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 3.0 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 3.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0061700 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.4 | 5.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.1 | 3.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.0 | 3.0 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 1.0 | 6.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.0 | 5.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.9 | 10.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.9 | 4.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.8 | 5.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.8 | 5.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.8 | 14.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.8 | 9.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.7 | 0.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.7 | 2.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 6.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.7 | 26.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.7 | 2.7 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.7 | 3.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.7 | 12.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.7 | 11.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.6 | 3.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.6 | 3.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.6 | 3.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 1.8 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.6 | 1.7 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.6 | 7.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.6 | 3.9 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.6 | 3.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 1.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.5 | 2.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 3.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.5 | 3.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 1.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.5 | 1.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.5 | 3.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.5 | 2.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 3.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 1.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 1.5 | GO:0046911 | hydroxyapatite binding(GO:0046848) metal chelating activity(GO:0046911) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.5 | 1.5 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.5 | 2.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 3.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 2.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.4 | 10.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 1.3 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.4 | 5.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 3.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 1.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 1.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 1.2 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.4 | 1.1 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.4 | 1.1 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.4 | 1.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 4.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.4 | 9.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 3.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.4 | 1.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 3.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.0 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.3 | 1.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.3 | 3.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 11.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 1.0 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 6.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 0.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 9.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 1.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 0.9 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.3 | 11.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 0.9 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 1.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 1.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.3 | 12.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 0.9 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.3 | 4.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 2.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 2.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 1.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 2.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 2.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 2.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 0.8 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.3 | 1.0 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.3 | 5.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 4.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 2.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 3.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 1.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 2.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 3.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 0.7 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.1 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 3.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.7 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.2 | 0.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 5.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 0.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 2.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 0.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 0.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 0.6 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 6.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 14.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 6.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.2 | 1.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 1.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 3.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 0.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 2.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 5.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 0.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 0.9 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.2 | 0.9 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.2 | 2.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 0.7 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 1.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 5.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 1.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 3.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 7.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.5 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.2 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 1.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 0.5 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 2.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 1.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.6 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.5 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.2 | 1.6 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 1.6 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 1.7 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 5.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 0.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 4.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 13.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.9 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 3.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 5.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 4.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 2.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.7 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.1 | 0.5 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 2.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.5 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.9 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.6 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 2.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 1.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.3 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 2.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0019863 | IgE binding(GO:0019863) Fc-gamma receptor I complex binding(GO:0034988) oligosaccharide binding(GO:0070492) |
| 0.1 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.6 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.1 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.3 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 1.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.9 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.9 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 6.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 2.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.8 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 3.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.6 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 4.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 1.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 1.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 2.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 0.4 | GO:0005168 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 2.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 2.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.4 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.5 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 1.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 10.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 19.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 4.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 5.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 1.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0015464 | acetylcholine receptor activity(GO:0015464) acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0032813 | death receptor binding(GO:0005123) tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 1.7 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 5.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.4 | 17.3 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.3 | 11.2 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 12.4 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 14.7 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.2 | 4.9 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.2 | 10.9 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 6.4 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 28.8 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 8.3 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.2 | 7.5 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.2 | 9.9 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.2 | 1.8 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.2 | 5.2 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 6.2 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.2 | 8.1 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.2 | 4.6 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.4 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.2 | 5.0 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 4.4 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.3 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 2.7 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.7 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.3 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 2.0 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.4 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.9 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.1 | 3.4 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.0 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.5 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.6 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.3 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.1 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.0 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.9 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.5 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 3.5 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.2 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.9 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.5 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 1.1 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.6 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 4.0 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.7 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 0.7 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.8 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.4 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.6 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.6 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 1.0 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.0 | 0.1 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.7 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 10.4 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 2.7 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.5 | 3.2 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 7.0 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 5.8 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.4 | 2.9 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 9.5 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 11.4 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.4 | 5.2 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 6.3 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 9.0 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 3.4 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 13.2 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 4.6 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 12.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 5.4 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 5.7 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 0.6 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 0.5 | REACTOME_PD1_SIGNALING | Genes involved in PD-1 signaling |
| 0.3 | 2.3 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.2 | 7.1 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.2 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 8.9 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 4.8 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 1.6 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 7.7 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 4.5 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 5.8 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.0 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.3 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 3.6 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 2.4 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 1.6 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 0.5 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.2 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 5.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.8 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.7 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.9 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.0 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.5 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.1 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.1 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.2 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.7 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.0 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.2 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 3.4 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.0 | REACTOME_THE_ROLE_OF_NEF_IN_HIV1_REPLICATION_AND_DISEASE_PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.9 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.5 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.6 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.2 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.4 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 0.8 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.1 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.6 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.6 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.5 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.9 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.5 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.2 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.8 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.5 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.2 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.1 | REACTOME_DOUBLE_STRAND_BREAK_REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 0.4 | REACTOME_DAG_AND_IP3_SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.9 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 6.3 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.9 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.5 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.0 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.1 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 0.7 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.1 | 0.3 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.2 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.6 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.2 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.7 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.1 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.6 | REACTOME_CTLA4_INHIBITORY_SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.8 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME_CHONDROITIN_SULFATE_DERMATAN_SULFATE_METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.3 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.7 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.1 | REACTOME_REGULATION_OF_INSULIN_SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.6 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 3.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 1.1 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.1 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.6 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.3 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.6 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME_FATTY_ACID_TRIACYLGLYCEROL_AND_KETONE_BODY_METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.1 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |